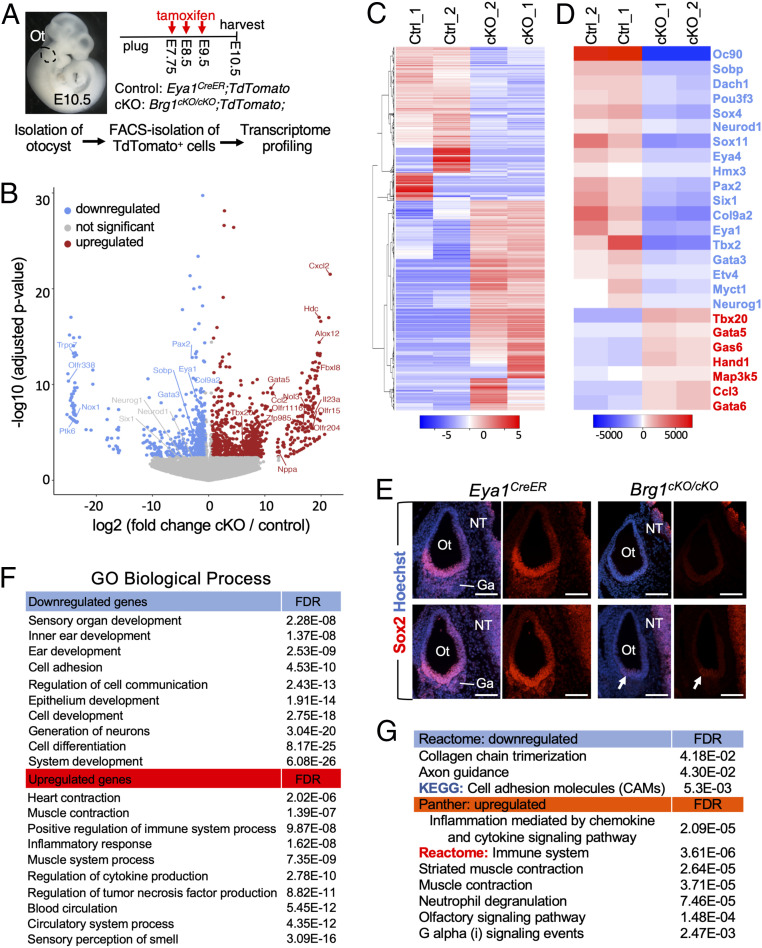
Fig. 3.
Transcriptome analysis reveals the requirement of Brg1 for specifying otic neurosensory lineage and a repressive function of nonotic gene expression. (A) Schematic drawing of TdTomato+ cell isolation. Otocysts (n = 8) were isolated from control and Brg1cKO/cKO littermates (n = 4 for each genotype harvested from 3 pregnant females) and dissociated into single cells. Five thousand FACS-sorted TdTomato+ cells were used for each biological replicate (samples “1” and “2”). Magnification, 6×. (B) Volcano plot showing transcripts differentially responding to depletion of Brg1. (C and D) Heatmap showing expression of all 3,092 (C) or 25 selected (D) differentially expressed genes. Blue or red indicates down- or up-regulated genes in Brg1cKO/cKO. Twenty-five selected genes include otic genes (in blue) known to be important for neurosensory progenitor development and TFs critical for heart as well as apoptotic and inflammatory proteins (in red). (E) Anti-Sox2 immunostaining on sections at E10.5. Arrows point to weak Sox2 signal in a few cells in the ventral otocyst (Ot). No Sox2+ progenitors in VII–VIIIth ganglionic region (Ga) were detectable in Brg1cKO/cKO, but Sox2 expression in the neural tube (NT) is unaffected. Scale bars, 100 μm. (F and G) GO (F) and pathway (G) enrichment analysis for differentially expressed genes. Blue or red indicates down- or up-regulated genes. GO, Panther and reactome pathway annotation tools (geneontology.org), heatmap and KEGG pathway tools (bioinformatics.sdstate.edu).