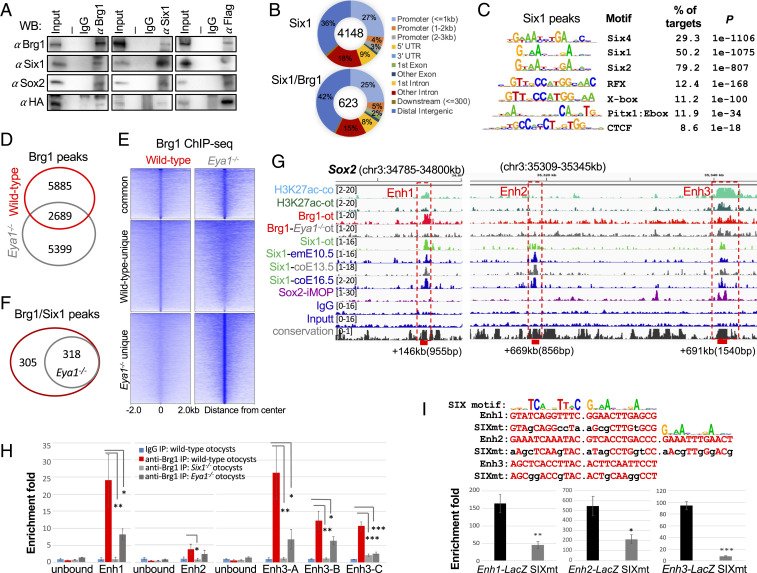
Fig. 5.
Brg1 physically forms protein complex with Eya1/Six1 and Brg1 enrichment at distal 3′ Sox2 enhancer regions is disrupted in the absence of Eya1 or Six1. (A) CoIP analysis using nuclear extracts prepared from E10.5 otocysts of Eya1HA-Flag embryos. Anti-Brg1, -Flag, or -Six1 was used for IP and antibodies used for Western blot are indicated. Input was 5% of the amount used for IP. Row Western blots are shown in SI Appendix, Fig. S5A. (B) Pie charts showing genomic distribution of Six1-binding sites and its overlapping peaks with Brg1. (C) Sequence logos of the significantly enriched top motifs from Homer Known motif analysis, letter size indicates nucleotide frequency. Percent of target sites in Brg1 peaks with significance of motif occurrence (P value) are indicated. (D) Venn diagram indicating overlap of Brg1 sites in wild-type and Eya1−/− otocyst. (E) Heatmaps of Brg1 peaks shown in D within a −2-kb/+2-kb window centered on Brg1 peaks. (F) Venn diagram indicating overlap of sites cooccupied by Brg1/Six1. (G) Genome browser visualization of overlapping occupancy of H3K27ac, Brg1, Six1, and Sox2 to the three distal 3′ Sox2 enhancer elements (boxes). Samples used for comparison: H3K27ac-co, E13.5 cochleae; H3K27ac-ot, E10.5 otocysts (ot); Brg1-ot, E10.5 otocysts; Brg1-Eya1−/−-ot, Eya1−/− otocysts; Six1-ot, E10.5 otocysts; Six1-emE10.5, E10.5 embryos; Six1-coE13.5 or -coE16.5, E13.5 or E16.5 cochleae; Sox2-iMOP, immortalized iMOP. (H) ChIP-qPCR confirms Brg1-binding to the Enh1 to -3 regions in E10.5 otocyst and a large decrease in Brg1-enrichment in Eya1−/− or Six1−/− otocyst. Primers for two Brg1 unbound regions as negative controls and three different primer sets (SI Appendix, Table S3) within the Enh3 region (”A,” “B,” and “C”) were used for qPCR. qPCR was performed in triplicates and the enrichment of mock IP (IgG) was considered onefold. *P < 0.05, **P < 0.01, and ***P < 0.001 determined by Student’s t test. (I) Sox2 Enh1 and Enh3 contain two SIX motifs, while Enh2 has three SIX motifs. Mutations introduced into the predicted SIX-motifs for each enhancer is indicated. Six1-binding to these reporters were assessed by ChIP-qPCR using chromatin prepared from 293 cells cotransfected with His-Six1 expression plasmid and each reporter, respectively. The enrichment of mock IP was considered onefold. *P < 0.05, **P < 0.01, and ***P < 0.001 determined by Student’s t test.