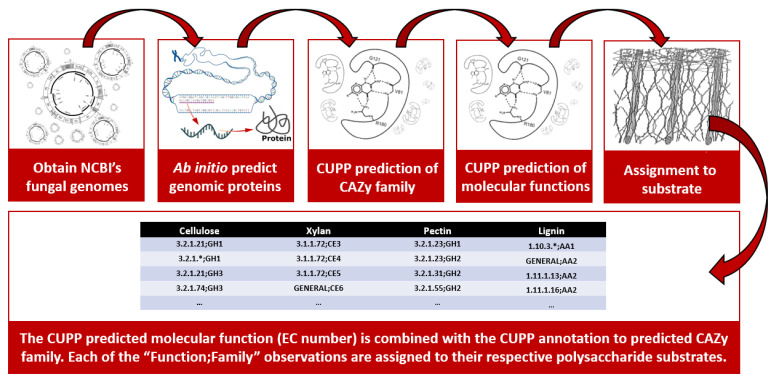
Figure 1.
Flow diagram describing the steps from downloading the full fungal genomes from NCBI and prediction of genomic proteins using AUGUSTUS. The carbohydrate-active enzymes were assigned a CAZy family and molecular function prediction via CUPP (see text). The predicted Function;Family observations were then assigned to their respective target substrates. The * indication added as fourht number in some EC numbers (molecular function numbers) indicate that no full EC number is available in the CAZy database. A similar molecular function (same EC number) from two different CAZy families are considered as two different observations by combining the EC number and the CAZy family into a combined string.