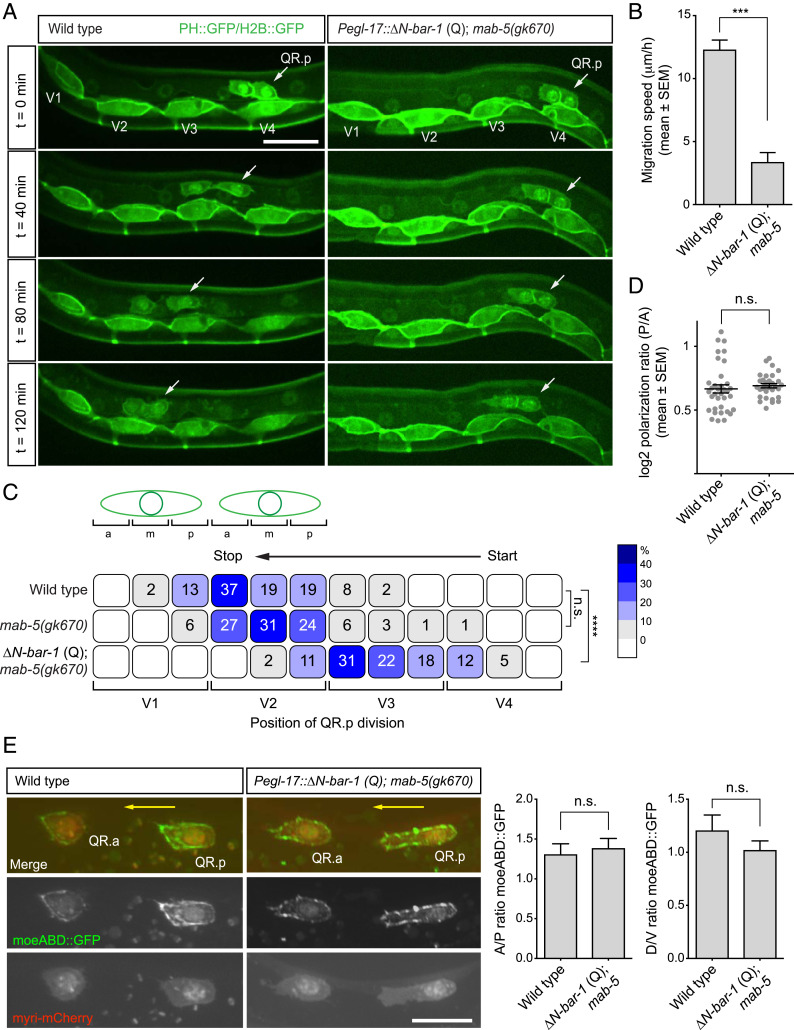
Fig. 1.
Activation of canonical Wnt signaling inhibits QR.p migration. (A) Time-lapse imaging of QR.p migration in wild type and animals specifically expressing ΔN-BAR-1 in the Q lineage (ΔN-BAR-1 Q). The seam (V) cells and QR.a and QR.p are marked with nuclear (H2B) and membrane-localized (PH) GFP (huIs63) (38). Anterior is left and dorsal is up (scale bar, 15 μm). (B) The average speed of QR.p during the first hour of migration. The bars represent mean ± SEM (n > 50 for all genotypes). The statistical significance was calculated using an unpaired t test (***P < 0.001). (C) The position of QR.p division with respect to the seam cells. Position at the anterior (a), middle (m), or posterior (p) side of the seam cell are indicated as percentiles of the total number of cells analyzed (n > 50 for all genotypes). The statistical significance was calculated using Fisher’s exact test (n.s., P ≥ 0.05, ****P < 0.0001). (D) Quantification of QR.p polarity as calculated by the ratio of the distance from the nucleus to the posterior and the anterior side of the cell. The black lines indicate mean ± SEM (n > 30 for all genotypes). The statistical significance was calculated using an unpaired t test (n.s., P ≥ 0.05). (E) Representative images of filamentous actin (F-actin) localization in QR.p in wild type and ΔN-BAR-1 (Q)–expressing animals. A moesin actin binding domain fused to GFP (moeABD::GFP) was used to visualize F-actin (44) and myristoylated mCherry as a marker for the Q cells. The ratio of moeABD::GFP along the anteroposterior and dorsoventral axes was quantified. The bars represent mean ± SEM (n > 20 for all genotypes) (scale bar, 10 μm). The statistical significance was calculated using an unpaired t test (n.s., P ≥ 0.05). The ΔN-BAR-1 (Q)–expressing strains contain mab-5(gk670).