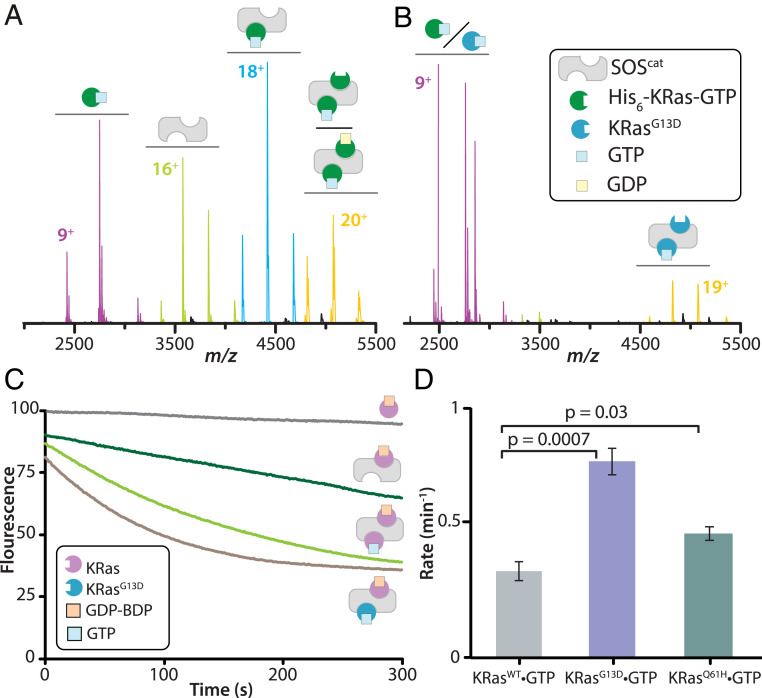
Significance
Disrupting the interaction between Ras and SOS has emerged as an attractive therapeutic strategy. Here, we characterized the assembly of the catalytic domain of SOS (SOScat) with oncogenic KRas mutants using native mass spectrometry. By resolving distinct molecular species, we show KRas mutants engage SOScat differently and the G13D KRas mutant robustly engages SOScat. A structure of KRasG13D in complex with SOScat shows repositioning of switch I and II regions within KRasG13D. Moreover, potent small-molecule Ras•SOS disruptors do not dissociate KRasG13D•SOScat complexes when KRasG13D–GTP is bound at allosteric site of SOScat. These results underscore the need for more potent Ras•SOS disruptors when considering targeting the disruption of different oncogenic Ras mutants in complex with SOS.
Keywords: cancer, Ras proteins, native mass spectrometry, Ras-SOS
Abstract
Ras is regulated by a specific guanine nucleotide exchange factor Son of Sevenless (SOS), which facilitates the exchange of inactive, GDP-bound Ras with GTP. The catalytic activity of SOS is also allosterically modulated by an active Ras (Ras–GTP). However, it remains poorly understood how oncogenic Ras mutants interact with SOS and modulate its activity. Here, native ion mobility–mass spectrometry is employed to monitor the assembly of the catalytic domain of SOS (SOScat) with KRas and three cancer-associated mutants (G12C, G13D, and Q61H), leading to the discovery of different molecular assemblies and distinct conformers of SOScat engaging KRas. We also find KRasG13D exhibits high affinity for SOScat and is a potent allosteric modulator of its activity. A structure of the KRasG13D•SOScat complex was determined using cryogenic electron microscopy providing insight into the enhanced affinity of the mutant protein. In addition, we find that KRasG13D–GTP can allosterically increase the nucleotide exchange rate of KRas at the active site more than twofold compared to KRas–GTP. Furthermore, small-molecule Ras•SOS disruptors fail to dissociate KRasG13D•SOScat complexes, underscoring the need for more potent disruptors. Taken together, a better understanding of the interaction between oncogenic Ras mutants and SOS will provide avenues for improved therapeutic interventions.
Ras, a member of the small G-protein family, represents important signaling molecules with diverse cellular roles, such as cell differentiation and proliferation (1–4). Different isoforms of Ras (HRas, KRas, and NRas) have high overall sequence identity and are the most commonly mutated of all discovered oncogenes (5, 6). Of the three Ras isoforms, KRas is the most frequently mutated isoform in cancers, such as pancreatic cancer (70–90%), colon cancer (30–50%), and lung cancer (20–30%) (7, 8). Ras proteins regulate cell signaling pathways by cycling between inactive, GDP-bound, and active, GTP-bound states that is accompanied by remodeling of three key regions within Ras: p-loop (residues 10–17), switch I (residues 30–38), and switch II (residues 60–76) (9–11).
As Ras proteins possess slow guanine nucleotide exchange rates, their activation is regulated by guanine nucleotide exchange factors (GEFs) that reload Ras with GTP (12, 13). The multidomain protein, Son of Sevenless (SOS), is a GEF with the cdc25 and Ras exchanger motif domains representing the minimal, functionally competent unit, termed SOScat (14). Structural studies have revealed two Ras binding sites to SOScat, leading to the discovery that binding of Ras–GTP at the distal (or allosteric) site allosterically modulates SOScat activity, which markedly increases the nucleotide exchange rate at the active site (14, 15). In addition, the degree of this allosteric modulation greatly depends on the nucleotide-bound state of Ras (14, 16, 17). Moreover, SOS is conformationally dynamic and binding of Ras at the allosteric site appears to shift the population to active conformation(s) of SOS (18). In addition, SOScat samples a broad range of turnover rates by fluctuating between distinct, long-lived functional states (19). Despite these advances, the nucleotide specificity of Ras bound to the active and allosteric sites of SOS and assembly with RAS, including oncogenic mutants, at the molecular level is poorly understood.
Targeting oncogenic Ras mutants presents significant challenges because of their relatively smooth surface that lacks potentially druggable pockets (6). Nevertheless, the discovery of KRas inhibitors, particularly those that form irreversible covalent bonds with Cys-12, comprises one of the most active areas of cancer research (6, 20). As there are few windows of opportunity to specifically target Ras mutants (6), apart from covalent binding to Cys-12, other approaches have also been explored, such as designing molecules to disrupt Ras•SOS interactions, thereby preventing activation of Ras (20). An increasing number of small-molecule disruptors and peptide mimetics have been designed to disrupt the Ras•SOS interaction (21). Potent small-molecule disruptors have recently been discovered that inhibit the formation of the Ras•SOS complex and demonstrate antiproliferative activity, representing a viable approach for targeting Ras-driven tumors (22, 23). These results highlight the therapeutic importance of disrupting the interaction between Ras and SOS.
Over the past three decades, native ion mobility–mass spectrometry (IM-MS) has evolved as a powerful analytical technique to investigate protein complexes and their interaction with other molecules (24–26). In native MS, biological samples in aqueous ammonium acetate are ionized using nanoelectrospray ionization and introduced into a mass spectrometer tuned to preserve noncovalent interactions and structure (24, 27). Native IM-MS can provide information on protein complexes, such as subunit stoichiometry and topology (28), and, unlike other biophysical techniques, resolve individual ligand-binding events (29). In combination with an apparatus to control temperature, native MS can be used to determine equilibrium binding constants and thermodynamics for protein–ligand and protein–protein that are in direct agreement with traditional biophysical approaches, such as isothermal calorimetry (30–35). Recently, native MS has been employed to determine transition state thermodynamics for the intrinsic GTPase activity of KRas and several oncogenic mutants (36). Notably, intrinsic GTPase activity rates determined using traditional solution-based assays mirrored those obtained using native MS (36).
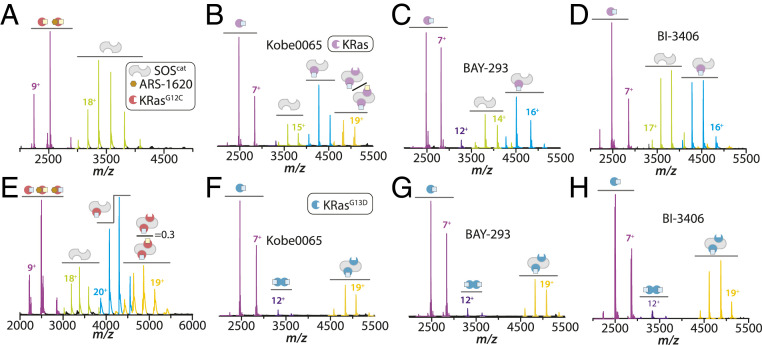
Although the interaction of Ras and SOS has been the subject of numerous studies, the interaction of oncogenic Ras mutants with SOS remains poorly described. To better understand the role of these interactions in cancer, native IM-MS is used to characterize the molecular assemblies formed between SOScat and mutants of KRas associated with cancer. IM spectrometry shows conformational heterogeneity of SOScat with specific conformers engaging KRas. Three selected oncogenic mutants of KRas form distinct molecular assemblies with SOScat, such as KRasG13D forming exclusively a ternary complex with SOScat. The cryogenic electron microscopy (cryo-EM) structure of the KRasG13D•SOScat complex provides insight into the mechanism for the higher affinity of KRasG13D for SOScat. KRasG13D–GTP also allosterically modulates the activity of SOScat more than the wild-type protein. In addition, the recent inhibitors developed to disrupt the KRas•SOScat complex, BAY-293 and BI-3406, cannot dissociate KRasG13D•SOScat complexes. Other small molecules, such as ARS-1620 and Kobee0065, display a range of efficacies in disrupting complexes formed between KRas mutants and SOScat.
Results
Conformational Dynamics of SOScat and Conformational Selection of KRas.
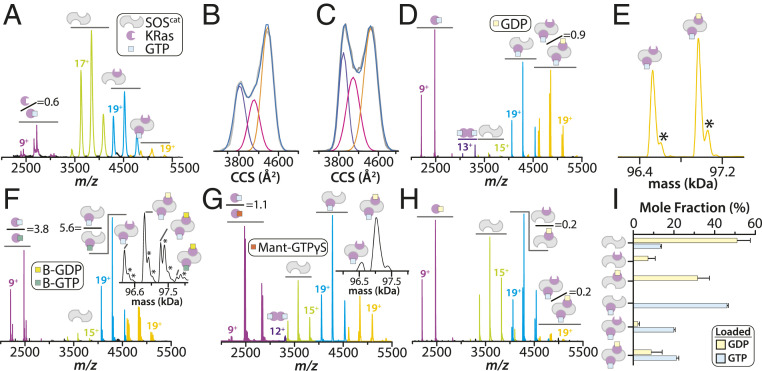
To investigate the interaction between SOS and Ras, we selected SOScat, which can bind up to two Ras molecules, and KRas due to its importance in cancer (7, 8). The KRas•SOScat complex purified following methods established for structural studies (14, 15) was subjected to native IM-MS analysis, a biophysical technique that has recently monitored the intrinsic GTPase activity of KRas (36). The native mass spectrum of the purified complex revealed an equilibrium of molecular species (Fig. 1A). The KRas•SOScat complexes contained a nucleotide-free KRas and the ternary complex, KRas•SOScat•KRas–GTP–Mg2+ (active site•SOScat•allosteric site), composed of one KRas and KRas bound to GTP. Based on previous NMR and spectroscopic results showing the low affinity of KRas–GTP to the active site (15, 16), we presume KRas–GTP is bound to the allosteric site of SOScat. The stoichiometry of this ternary complex is consistent with crystal structures (14). Ion mobility measurements, which report on the rotationally averaged collision cross-section (CCS), show the existence of different conformers populated by SOScat, which could be modeled with a minimum of three different Gaussian distributions (Fig. 1B and SI Appendix, Fig. S1 and Table S1). A similar analysis of SOScat but in the absence of KRas (Fig. 1C and SI Appendix, Fig. S1) revealed a marked increase in the most compact conformers, which are depleted when KRas is present (Fig. 1B). The lack of SOScat repopulating these conformers in the presence of KRas is entirely consistent with the reported long-lived interconverting dynamical states of SOScat (18, 19). In another words, the observed depletion in SOScat conformers that preferentially engage KRas could only happen if the different conformers have slow interconverting rates and long-lived states, which do not repopulate or equilibrate on the timescale of the experiment, hence their observed depletion in abundance. These findings provide additional evidence of conformational dynamics of SOScat and conformational selection for binding KRas.
Fig. 1.
Conformational dynamics and molecular assemblies of SOScat and KRas. (A) Native mass spectrum of 2 µM KRas•SOScat complex purified by size exclusion chromatography. Mass spectral peaks corresponding to KRas, SOScat, binary, and ternary complexes are colored purple, chartreuse, cyan, and orange, respectively. (B) Collision cross-section (CCS) distribution for the 16+ ion of SOScat (blue lines). Regression (R2 = 1.0) of three Gaussian peaks (purple, pink, and orange lines) and their sum (gray line). (C) CCS profile for the 16+ ion of SOScat recorded in the absence of KRas. Shown as described in B. (D) Mass spectrum of 2 µM SOScat and 6 µM KRas–GTP recorded immediately after mixing. (E) Deconvolution of mass spectrum in D and selected mass range corresponding to ternary complexes. Inorganic phosphate adducts are denoted by an asterisk. (F and G) Mass spectrum for the mixture described in D with the addition of (F) 2 µM BODIPY modified GTP (B-GTP) and (G) 5 µM Mant-GTPS. (H) Mass spectrum of 2 µM SOScat and 6 µM KRas–GDP recorded immediately after mixing. (I) Plot of the mole fraction of SOScat complexes determined from deconvolution of mass spectra. Reported are the mean and SD (n = 3). Mass spectra shown in D–H were acquired on an Exactive plus EMR Orbitrap mass analyzer. KRas bound at active and allosteric sites are shown at Top Right and Bottom Left of the cartoon, respectively.
Molecular Assemblies of KRas–GTP and SOScat.
To better understand the molecular assemblies of KRas with SOScat in the presence of unmodified nucleotides, we conducted studies using a higher-resolution mass spectrometer (29). We first mixed SOScat with a threefold molar excess of KRas loaded with GTP (KRas–GTP) (SI Appendix, Fig. S2). The mass spectrum recorded immediately after mixing shows the presence of monomeric and dimeric KRas bound to GTP, and a single binary complex composed of SOScat•KRas–GTP–Mg2+ (Fig. 1 D and I). In addition, two ternary complexes of near equal abundance were measured corresponding to KRas•SOScat•KRas–GTP–Mg2+(GDP)0–1 (Fig. 1E). The measured molecular weight of these complexes and of the proteins alone are in good agreement with theoretical values (SI Appendix, Table S2). Notably, these molecular species were not resolved in the lower-resolution mass spectrum (Fig. 1A). KRas bound to GTP dominates the binary and ternary complexes, a result that is consistent with the higher affinity of KRas–GTP to the allosteric site (15, 16). These findings capture an equilibrium of molecular assemblies formed between KRas and SOScat that would be difficult to render using other biophysical techniques.
SOScat Stimulates KRas GTPase Activity.
Despite addition of KRas–GTP to SOScat, an additional peak corresponding to KRas•SOScat •KRas–GTP–Mg2+(GDP)1 was detected with a measured mass (96,972.2 ± 2.3 Da) in close agreement with the calculated mass (96,964.8 Da) (SI Appendix, Table S2). Mass selection of this ternary complex in the quadrupole followed by collision-induced dissociation (CID) further corroborates this assignment (SI Appendix, Fig. S3). In addition, surface-induced dissociation (SID) of the isolated ternary complex resulted in dissociation of KRas and KRas–GTP from the complex (SI Appendix, Fig. S4). Notably, KRas was loaded with GTP (∼98%) (SI Appendix, Fig. S2) and the slow intrinsic GTP hydrolysis of KRas in the absence of SOScat cannot account for the perplexing abundance of GDP. The appearance of GDP is most likely due to the GTPase activity of KRas but stimulated by SOScat, which would result in production of GDP and inorganic phosphate (H2PO4−). In crystal structures (14), phosphate is bound to ternary complexes, and accordingly here phosphate adducts are observed on the complex with a measured mass of 97,060.2 ± 2.1 Da compared to the theoretical mass of 97,061.8 Da (Fig. 1E). Moreover, incubation of a mixture of KRas–GTP and SOScat overnight resulted in nearly twofold greater levels of KRas bound to GDP compared to the control solution containing only KRas–GTP (SI Appendix, Fig. S5). An inorganic phosphate assay also corroborated these findings with higher phosphate concentration when KRas–GTP is in the presence of SOScat compared to the control KRas–GTP solution (SI Appendix, Table S3). Taken together, these results indicate a role of SOS in stimulating the GTPase activity of KRas.
Additional evidence for stimulation of KRas GTPase activity is offered by the use of GTP analogs. First, a BODIPY-modified GTP (B-GTP) analog was added to an incubated mixture of KRas–GTP and SOScat followed by recording the native mass spectrum. Multiple species are observed along with adducts corresponding to inorganic phosphate and B-GDP only bound to KRas in complex with SOScat (Fig. 1F). Interestingly, the lower abundance of KRas•SOScat •KRas–B-GTP⋅Mg2+(B-GDP)1 compared to KRas•SOScat •KRas–GTP–Mg2+(B-GDP)1 suggests the allosteric binding site is less dynamic than the active site (Fig. 1 F, Inset). Next, the nonhydrolyzable GTP analog, Mant-GTPS, was added to the incubated mixture of SOScat and KRas–GTP. The native mass spectrum shows no detectable signal corresponding to hydrolyzed Mant-GDP, and Mant-GTPS is only bound to free KRas (Fig. 1G). These findings show various molecular assemblies of KRas and SOScat but also report that SOScat can stimulate of KRas GTPase activity, a characteristic feature of GTPase-activating proteins (GAPs) (4).
Molecular Assemblies of KRas–GDP and SOScat.
As the degree of allosteric modulation of SOScat greatly depends on the nucleotide-bound state of Ras (14, 16), KRas was loaded with GDP (KRas–GDP) prior to mixing with SOScat. The native mass spectrum for this mixture is dominated by two binary complexes with the majority (∼80%) bound to GDP (Fig. 1 H and I). In addition, there are low abundant signals corresponding to KRas•SOScat •KRas–GTP–Mg2+(GDP)0–1 that stem from a small fraction of KRas–GTP not loaded with GDP (SI Appendix, Fig. S6), underscoring the sensitivity of our measurements. KRas–GTP promotes the formation of a ternary complex, which is in accord with previous studies (14, 16).
Molecular Assemblies of SOScat and KRas Oncogenic Mutants.
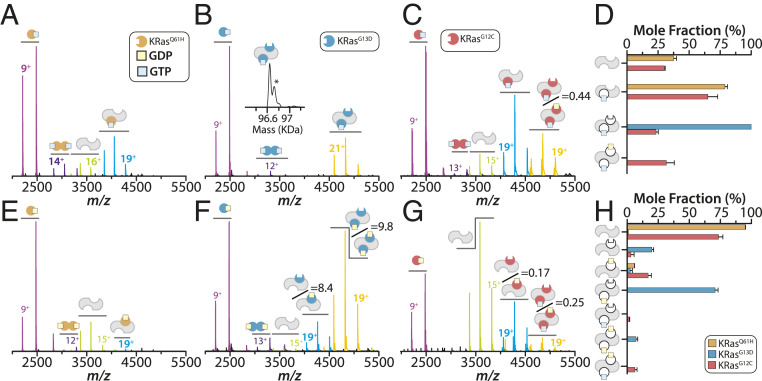
Three oncogenic mutants of KRas were selected based on their high occurrence in some cancers (Q61H, G13D, and G12C) (5). The mutant proteins were first loaded with GTP and mixed with SOScat immediately before native MS analysis. The mass spectrum for KRasQ61H and SOScat mixture showed a higher abundance of dimeric KRasQ61H with each bound to GTP and no detectable ternary complex (Fig. 2 A and D). The SOScat•KRasQ61H–GTP–Mg2+ complex uniformly contained one GTP and no ternary complexes were observed (Fig. 2 A and D). In stark contrast, KRasG13D–GTP solely formed a KRasG13D•SOScat •KRasG13D–GTP–Mg2+ complex (Fig. 2 B and D). The third GTP-loaded mutant KRasG12C engaged SOScat in a similar fashion as wild-type KRas with the exception of an increased abundance of the ternary complex containing GDP (Fig. 2 C and D and SI Appendix, Fig. S7). Of the three oncogenic mutants, GDP was observed only in SOScat•KRasG12C complexes. In summary, oncogenic mutants form assemblies with SOScat that are distinct from those formed with wild-type KRas.
Fig. 2.
Distinct molecular assemblies of SOScat with oncogenic KRas mutants. (A–C) Native mass spectra of 2 µM SOScat mixed with three equivalents of (A) KRasQ61H–GTP, (B) KRasG13D–GTP, or (C) KRasG12C–GTP. Mass spectra are shown as described in Fig. 1. (D) Plot of the mole fraction of SOScat complexes formed with GTP-loaded proteins. (E–G) Mass spectra of 2 µM SOScat mixed with threefold molar excess of (E) KRasQ61H–GDP, (F) KRasG13D–GDP, or (G) KRasG12C–GDP. (H) Plot of the mole fraction of SOScat complexes formed with GDP-loaded proteins. Shown as described in Fig. 1.
We next investigated the three oncogenic mutants loaded with GDP and their assembly with SOScat. The mass spectrum of a 3:1 mixture of KRasQ61H to SOScat had weak signal for a GDP-bound binary complex (Fig. 2 E and H). KRasG13D–GDP predominantly formed ternary complexes composed of KRasG13D•SOScat•KRasG13D–GDP(GDP)0–1 with ∼90% of the signal accounting for the complex bound to only one GDP (Fig. 2 F and H). This unexpected observation of a prevalent ternary complex for KRasG13D–GDP suggests it can bind the allosteric site and possibly function as an allosteric modulator of SOScat. Assembly of SOScat and KRasG12C–GDP led to the formation of complexes reminiscent of wild-type KRas but overall lower in abundance (Fig. 2 G and H).
Complexes of KRas with a Mutant Form of SOScat.
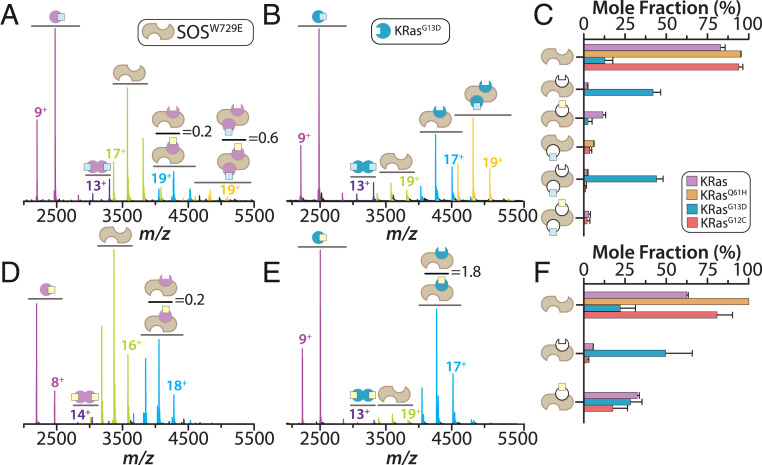
SOScat containing the W729E mutation (SOSW729E) has been reported to abolish Ras binding at the allosteric site (15). The mixture of KRas–GTP and SOSW729E showed a significant depletion of higher-order complexes (Fig. 3 A and C). Again, GDP is present in the ternary complex with a ratio skewed to those containing GDP. KRasQ61H–GTP and KRasG12C–GTP also displayed an overall reduction in complex formation (Fig. 3C and SI Appendix, Fig. S8). Surprisingly, KRasG13D–GTP formed complexes composed of KRasG13D•SOSW729E and KRasG13D•SOSW729E•KRasG13D–GTP–Mg2+ (Fig. 3 B and C). Moreover, SOSW729E and KRas–GDP formed two binary complexes, KRas•SOSW729E(GDP)0–1, in the same ratio but lower in signal abundance compared to SOScat alone (Fig. 3 D and F). KRasG13D–GDP primarily assembled into binary complexes with a majority (∼64%) containing nucleotide-free KRasG13D (Fig. 3 E and F). KRasG12C•SOSW729E(GDP)0–1 complexes were observed for the assembly of KRasG12C–GDP and SOSW729E, and KRasQ61H–GDP did not engage SOSW729E (Fig. 3F and SI Appendix, Fig. S8). Taken together, these results demonstrate a clear preference for the active site of SOScat toward KRas–GDP.
Fig. 3.
Complexes of a mutant form of SOScat with KRas and oncogenic mutants. (A and B) Native mass spectra of 2 µM SOSW729E mixed with 6 µM of (A) KRas–GTP or (B) KRasG13D–GTP. Mass spectra are shown as described in Fig. 1. (C) Plot of the mole fraction of SOSW729E complexes formed with GTP-loaded proteins. Shown as described in Fig. 1. (D and E) Mass spectra for a 1:3 mixture of SOSW729E with (D) KRas–GDP or (E) KRasG13D–GDP. (F) Plot of the mole fraction of SOSW729E complexes formed with GDP-loaded proteins. Shown as described in Fig. 1.
Structural Characterization of the KRasG13D–GTP and SOScat Ternary Complex.
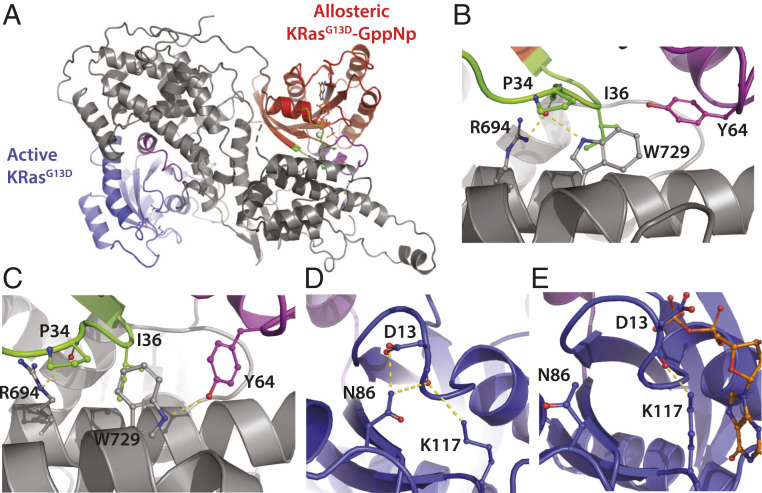
The unique properties of KRasG13D to form predominantly ternary complexes with SOScat prompted us to determine the structure using cryo-EM. The ternary complex was prepared using KRasG13D loaded with 5′-guanylyl imidodiphosphate (GppNp), a nonhydrolyzable analog of GTP (SI Appendix, Fig. S9). The structure of the complex was determined to a resolution of 3.47 Å (SI Appendix, Methods) with KRasG13D and KRasG13D–GTP bound at the active and allosteric sites of SOScat, respectively (Fig. 4A and SI Appendix, Fig. S10). The presence of nucleotide-free KRasG13D at the active site is in line with MS results (Fig. 2B). The structure is largely reminiscent with that of HRas•SOScat •HRas–GppNp (Protein Data Bank [PDB] 1NVW) with some notable structural differences, such as a ∼2.5-Å displacement of the KRasG13D molecule at the active site (SI Appendix, Fig. S11 E and F). Analysis of KRasG13D–GTP bound at the allosteric site reveals that KRasG13D adopts a state 2 conformation, characterized by interaction of the γ-phosphate with the side chain of Thr35 within switch I and the amide of Gly60 within switch II (SI Appendix, Fig. S11B) (37). Moreover, the structure of KRasG13D–GppNp bound at the allosteric site of SOScat aligns well with that of active conformation of HRas–GTP (SI Appendix, Fig. S11C) (37). However, the molecular interactions of KRasG13D–GTP bound at the allosteric site of SOScat differs from that observed for SOScat in complex with HRas–GTP. More specifically, the side chain of tryptophan at position 729 of SOScat is reoriented and interacting with the carbonyl of Pro34 of KRasG13D (Fig. 4 A and B). The orientation of Trp729 is similar to the crystal structure of the Tyr64Ala mutant of HRas bound at the allosteric site of SOScat (SI Appendix, Fig. S11D), which this mutant HRas protein has significantly reduced binding affinity to the active site of SOScat (38). Trp729 adopts a different orientation that interacts with Try64 in the structure of HRas–GppNp bound at the allosteric site of SOScat (Fig. 4C). The density for residues 62–67 within the switch II region was not clear, possibly due to this region populating different conformations. The most marked difference in the structure of the KRasG13D in complex with SOScat compared to other available structures (14, 22) is the structure of KRasG13D bound at the active site (Fig. 4D). Aspartic acid at position 13 forms hydrogen bonds with Asn86 and Lys117. These interactions, for example, reposition loop 8 by ∼4 Å compared to HRas in complex with SOScat along with displacing other regions, such as helices 3 and 4, p-loop, and switch I (Fig. 4D and SI Appendix, Fig. S11E). The observed conformation of KRasG13D also differs from the structure of a mutated form of KRasG12C bound only at the active site of SOScat (SI Appendix, Fig. S11F). Comparison of the KRasG13D–GDP structure (PDB 6E6G) with KRasG13D bound at the active site reveals the side chain of Asp13 is rotated 180° and pointing away from Asn86 (Fig. 4E).
Fig. 4.
Structure of KRasG13D in complex with SOScat. (A) The structure is shown in cartoon representation with KRasG13D molecules bound at the active and allosteric sites of SOScat (gray), which are colored blue and red, respectively. Switch I and II regions of KRasG13D are colored in green and purple, respectively. Allosteric KRasG13D is bound to GppNp and magnesium. (B) Molecular interactions formed at the interface between KRasG13D–GppNp and SOScat. Hydrogen bonds are shown as yellow dashed lines. (C) The allosteric interface for HRas–GppNp and SOScat (PDB 1NVW) (14) shown in similar orientation as B. (D) KRasG13D bound at the active site with Asp13 forming interactions with Asn86 and K117. (E) X-ray structure of KRasG13D bound to GDP (PDB 6E6G) (51) shows the side chain of Asp13 is oriented in the opposite direction, away from Asn86.
The Modulation of SOScat Activity by KRasG13D–GTP.
Competition and nucleotide exchange assays were conducted to better understand the significance of the interaction between KRasG13D and SOScat. First, the complex formed between SOScat with GTP-loaded KRas harboring the N-terminal hexahistidine affinity tag (His6–KRas–GTP) displayed similar abundances of molecular species as the tagless protein (Fig. 5A). Interestingly, addition of KRasG13D–GTP to this complex not only completely abolished the binding of wild-type KRas to SOScat but predominantly formed a ternary complex of KRasG13D•SOScat •KRasG13D–GTP–Mg2+ (Fig. 5B). The experiment performed in the opposite fashion, where His6–KRas–GTP is added to preformed complexes of KRasG13D–GTP and SOScat, resulted in no disruption of KRasG13D•SOScat complexes (SI Appendix, Fig. S12). Next, the SOScat-stimulated nucleotide exchange rate of KRas bound to B-GDP in the absence and presence of KRasG13D–GTP was determined. The intrinsic nucleotide exchange rate of KRas–B-GDP is slow (39) (k = 0.12 min−1) but enhanced in the presence of SOScat (Fig. 5C). The rate of nucleotide exchange is allosterically modulated (k = 0.3 min−1) in the presence of KRas–GTP (Fig. 5 C and D) consistent with previous studies (14). However, the addition of KRasG13D–GTP increased the exchange rate by more than twofold, k = 0.7 min−1 (Fig. 5 C and D). The addition of KRasQ61H–GTP (k = 0.4 min−1) marginally increased the nucleotide exchange rate compared to KRas–GTP and much less than KRasG13D–GTP. Attempts to measure SOScat-mediated nucleotide exchange rate of KRasG13D in the presence of allosteric KRasG13D–GTP was not possible due to extremely fast nucleotide exchange rate of this mutant protein consistent with previous studies (17). In short, KRasG13D exhibits not only a higher affinity for SOScat but is also a more potent allosteric modulator of SOScat activity.
Fig. 5.
Competition and cross-activation of KRasG13D–GTP. (A) Native mass spectrum of 1 µM SOScat in complex with 3 µM His6–KRas–GTP. (B) Native mass spectrum after adding KRasG13D–GTP to the mixture in A to a final concentration of 3 µM. Mass spectra are shown as described in Fig. 1. (C) Intrinsic (black line) and SOScat-mediated (dark green line) nucleotide exchange of KRas loaded with B-GDP. The addition of KRas–GTP (light green line) or KRasG13D–GTP (brown line) to a mixture of KRas–B-GDP and SOScat accelerates the nucleotide exchange rate. (D) Nucleotide exchange rates determined for data presented in C. The SOScat-mediated nucleotide exchange rate in the presence of KRasQ61H–GTP is also provided. Reported are the mean and SD from three independent experiments.
Disruption of the Interaction between KRas and SOScat.
As the disruption of the interaction between Ras and SOS is an attractive approach to curb aberrant Ras signaling (21, 40), we next investigated whether small-molecule Ras•SOS disruptors can efficiently disrupt complexes of SOScat and KRas mutants. Recent traction has been made on the development of specific covalent inhibitors of KRasG12C, such as ARS-1620 (20, 41). Reacting 10 equivalents of ARS-1620 with KRasG12C loaded with either GTP or GDP showed complete reactivity when the enzyme is bound to GDP and limited reactivity when GTP is bound (SI Appendix, Fig. S13). These results agree with the specificity of ARS-1620 toward the inactive, GDP-bound state of KRasG12C. Moreover, the addition of ARS-1620 to an incubated mixture of SOScat and KRasG12C–GDP resulted in complete disruption of complexes (Fig. 6A), consistent with a SOS-mediated nucleotide exchange assay (41). However, ARS-1620 added to a preincubated mixture of SOScat and KRasG12C–GTP had no appreciable disruption of binary and ternary complexes (Fig. 6E). This result suggests KRasG12C–GTP allosterically modulates the affinity of KRasG12C at the active site of SOScat, thereby hindering the reactivity of ARS-1620. These findings highlight the inability of ARS-1620 to disrupt assemblies of SOScat and KRasG12C–GTP.
Fig. 6.
The effect of ARS-1620, BAY-293, and Kobe0065 on assemblies of KRas and SOScat. (A–E) Mass spectrum recorded after adding 10 µM ARS-1620 to preincubated mixtures of 1 µM SOScat with 3 µM (A) KRasG12C–GDP and (E) KRASG12C–GTP. Mass spectra are shown as described in Fig. 1. (B–F) Mass spectrum recorded after adding 2.5 µM Kobe0065 to preincubated mixtures of 1 µM SOScat with 3 µM (B) KRas–GTP and (F) KRASG13D–GTP (C–G) Mass spectrum recorded after adding 2.5 µM BAY-293 to preincubated mixtures of 1 µM SOScat with 3 µM (C) KRas–GTP and (G) KRASG13D–GTP. (D–H) Mass spectrum recorded after adding 2.5 µM BI-3406 to preincubated mixtures of 1 µM SOScat with 3 µM (D) KRas–GTP and (H) KRASG13D–GTP.
We next tested the efficacy of small-molecule Ras•SOS disruptors, Kobe0065 (IC50 = 20 µM), BAY-293 (IC50 = 21 nM), and BI-3406 (IC50 = 5 nM) (22, 23, 42). Kobe0065 binds directly to Ras–GTP and is reported to alter the binding at the allosteric site of SOS (42). The addition of 2.5 equivalents of Kobe0065 to SOScat in complex with KRas–GTP or KRasG13D–GTP resulted in marginal and no disruption, respectively (Fig. 6 B and F). Increasing the concentration of Kobe0065 to 200 µM, 10 times the reported IC50 value, did not disrupt binary and ternary complexes. In addition, the small molecule was ineffective at disrupting complexes of KRasG12C and SOScat (SI Appendix, Fig. S14C). BAY-293 has been shown to efficiently disrupt the interaction between KRasG12C and SOScat leading to antiproliferative activity (22). However, the small molecule was not effective at disrupting the binding of KRas–GTP and KRasG12C–GTP at the allosteric site of SOScat (Fig. 6C and SI Appendix, Fig. S14B). BAY-293 binds directly to SOScat (SI Appendix, Fig. S14A) and disrupts the ternary complexes formed between SOScat and KRas or KRasG12C (Fig. 6C and SI Appendix, Fig. S14B). Moreover, BAY-293 did not disrupt the ternary complex of KRasG13D–GTP and SOScat (Fig. 6G). The compound, however, does inhibit the formation of complexes formed between SOSW729E and either KRas–GDP or KRasG13D–GDP (SI Appendix, Fig. S14 D and E). BI-3406 (23), a recently reported highly selective and potent inhibitor of SOS1, was screened for the ability to disrupt complexes formed between SOScat and wild-type and mutants of KRas. At 2.5 µM, BI-3406 binds directly to SOScat (SI Appendix, Fig. S15A) and disrupts the ternary complex between SOScat and KRas (Fig. 6D). In contrast, BI-3406 at the same concentration did not disrupt the ternary KRasG13D•SOScat complex (Fig. 6H), even after overnight incubation (SI Appendix, Fig. S15B). At higher concentrations of the compound (4,000-fold the reported IC50), the ternary complex between KRasG13D and SOScat was almost completely disrupted (SI Appendix, Fig. S15C). Similar to BAY-293, BI-3406 disrupts the binary complex between KRasG13D–GDP and SOSW729E (SI Appendix, Fig. S15D).
Discussion
Dynamics of signaling proteins play crucial roles in their function (43). Although conformational heterogeneity has been reported for SOScat (18, 19), ion mobility measurements provide direct experimental evidence of the protein populating at least three conformers in solution with distinct conformers engaging KRas. If the dynamics were fast, the depleted conformers that engage with KRas would be repopulated. Instead, depletion of specific conformers in the presence of KRas is in direct agreement with the long-lived, interconverting dynamical nature of SOScat (18, 19). This is a demonstration of ion mobility showing clear evidence of conformational selection for a protein–protein interaction.
Native MS provides a unique opportunity to monitor the molecular assemblies of SOScat with Ras. Unlike other approaches, the sensitivity and resolution of modern mass spectrometers enable not only measurements of proteins at low micromolar concentration but use of unmodified nucleotides. Native mass spectra resolve a number of molecular assemblies formed between KRas and SOScat that populate different nucleotide-bound states. The abundances of various nucleotide-bound states of binary and ternary complexes provide a glimpse into the affinity of nucleotides. For example, binary and ternary complexes of KRasG13D are dominated by nucleotide-free states that may have bearing to the reported fast nucleotide exchange rates of the mutant protein (39). We also find an unexpected abundance of GDP immediately after mixing KRas–GTP with SOScat, and addition of B-GTP to preformed ternary complex further validates the SOScat-mediated stimulation of KRas GTPase activity. However, no GDP was observed when a nonhydrolyzable analog is added to the complex. The presence of GDP was also observed for KRasG12C, which has reduced intrinsic GTPase activity compared to the wild-type protein (36). These results provide compelling evidence that SOScat has GAP-like characteristics. Nevertheless, it is unknown whether the hydrolysis reaction is carried out at the active or allosteric site(s) of SOScat. The observed ions corresponding to KRas•SOSW729E(GDP)1 after immediate MS analysis of a KRas–GTP and SOSW729E mixtures suggests that the hydrolysis reaction likely occurs at the active site. Furthermore, SOSW729E engineered to disrupt binding at the distal site shows a clear preference for binding KRas–GDP at the active site, consistent with previous reports (15–17). However, our data also show SOSW729E does not completely abolish binding at the distal site as reported (15), thereby convoluting the interpretation of reported binding affinities (15–17). In addition, signal is detected corresponding to dimers of wild type and oncogenic mutants of KRas. At the concentration of KRas (3 µM) used in the native MS studies, it is unlikely that the homodimers arise from nonspecific association and provide direct evidence of KRas dimerization, which has been implicated in Ras signaling (44).
The structure of KRasG13D•SOScat•KRasG13D–GppNp provides molecular details and clues to understanding the observed higher binding affinity of KRasG13D for SOScat. First, KRasG13D bound at the active site of SOScat is displaced compared to those in complex with HRas and KRasG12C (14, 22). Asp13 forms molecular interactions that reorients its side chain away from the nucleotide binding pocket that is essentially primed for binding GTP to a greater extent than the wild-type protein. This observation draws a corollary to native MS results, where the majority of the ternary complex is not bound to GDP. Aside from structural differences observed at the active site, the overall structural similarity to the HRas•SOScat•HRas–GppNp complex indicate the molecular interactions would be similar for HRas, KRas, and KRasG13D. Therefore, a plausible explanation for the observed higher binding affinity of KRasG13D is likely due to the altered dynamics of the mutant protein. In particular, switch II of KRasG13D is known to become less flexible, regardless of nucleotide-bound state, and positively correlated with β2–β3 loop motions (45). Importantly, switch II forms key interactions at the allosteric site of SOScat, and the altered dynamics of switch II in KRasG13D could result in an increase in the apparent binding affinity, i.e., a larger fraction of KRasG13D adopts a conformation(s) that can selectively engage SOScat. Moreover, the reduced dynamics of the switch II region of the KRasG13D–GDP can partly explain the ability of the GDP-bound protein to bind at the allosteric site and form ternary complexes with SOScat.
As oncogenic mutants have impaired GTPase activity, it has been surmised that activation of oncogenic Ras is independent of SOS (46). For example, activation of KRasG13D and KRasQ61H has been suggested to be independent of SOS due to their fast nucleotide exchange rate and impaired-GTPase activity, respectively (17, 39). However, results from native MS reveal KRas oncogenic mutants show an assortment of molecular assemblies ranging from weakly interacting KRasQ61H to KRasG13D that robustly engages SOScat. More specifically, KRasQ61H–GTP does not exert an allosteric effect implying that activation of this mutant is likely SOS independent. In contrast, KRasG13D–GTP unexpectedly forms predominantly a KRasG13D•SOScat•KRasG13D–GTP complex. KRasG13D–GDP also forms KRasG13D•SOScat •KRasG13D–GDP(GDP)0–1 complexes, suggesting the GDP-bound form of this mutant may serve as an allosteric modulator of SOScat. Moreover, KRasG13D–GTP is a potent allosteric modulator of SOScat and can also enhance the SOS-mediated nucleotide exchange of wild-type KRas. Considering cancer cells can be heterozygous for the KRasG13D mutation, these findings are of particular importance as SOS activity can be allosterically modulated by KRasG13D, regardless of nucleotide-bound state, which in turn would lead to robust SOS-mediated activation of KRas. These findings provide additional evidence that oncogenic Ras bound at the allosteric site of SOS can cross-activate KRas, which has been shown to be essential for tumorigenesis (47).
Ras activation is tightly regulated by SOS and disruption of this interaction is an attractive route for targeting Ras-driven cancers (21, 40). ARS-1620 (IC50, 120 nM) cannot disrupt assemblies of SOScat and KRasG12C–GTP, suggesting KRasG12C–GDP within the ternary complex is shielded from reaction with ARS-1620. KRasG12C–GTP allosterically promotes the interaction of KRasG12C at the active site, which diminishes the efficacy of the inhibitor to dissociate the complex. Kobe0065, designed to disrupt Ras binding at the allosteric site of SOScat, was ineffective for complexes formed with KRas, KRasG12C, and KRasG13D. The recent discovery of KRas•SOS disruptors have shown promising antiproliferative activity (22, 23). However, we show that while BAY-293 successfully disrupts KRas and KRasG12C complexes with SOScat, it fails to disrupt those formed with KRasG13D at 119-fold the IC50 (IC50 = 21 nM). Addition of BI-3406, 500-fold above the IC50 (IC50 = 5nM), did not disrupt the ternary complex of KRasG13D–GTP and SOScat. Although BAY-293 and BI-3406 compounds can disrupt KRasG13D binding to the active site of SOScat (SI Appendix, Figs. S14E and S15D), the presence of KRasG13D–GTP bound at the allosteric site of SOScat leads to enhanced stability of the ternary complex that is persistent to disruption with these small-molecule disruptors. The antitumor activity of BAY-293 and BI-3406 is greatly enhanced in combination with specific KRasG12C and MEK1 inhibitors (22, 23), and this strategy may mitigate cases where the efficacy of small-molecule Ras•SOS disruptors is reduced when SOS is engaged with oncogenic Ras mutants, such as KRasG13D. Overall, the ability of KRasG13D to exclusively form ternary complexes with SOScat, compete with KRas binding SOScat, activate SOScat to facilitate loading of wild-type KRas, and resistance to disruption by small-molecule disruptors may explain the aggressive biology of tumors associated with this mutant (48). Moreover, the higher binding affinity of KRasG13D, and likely other Ras mutants, for SOS may lead to not only an increase in the recruitment of SOS to the plasma membrane but also prolong dwell time allowing an increase in Ras activation by SOS (49). In closing, these results showcase the ability of native IM-MS to provide unprecedented insight into molecular assemblies, such as those formed between SOScat and KRas, and open avenues to develop more potent Ras-SOS inhibitors, especially for Ras mutants that robustly bind and activate SOS.
Methods
Protein Expression and Purification.
Human KRas4B (residues 1–169) and SOScat (residues 558–1049) were expressed and purified as previously described (36). The details of expression and purification are in SI Appendix, Materials and Methods. All mutations were generated using the Q5 site-directed mutagenesis kit (New England Biolabs) following the manufacturer’s protocol. Additional details are in SI Appendix, Materials and Methods.
Sample Preparation to Monitor the Assembly of the Complex via Native MS Analysis.
In order to obtain mass spectra of KRas or mutants (loaded with GDP or GTP) complexed with SOScat or SOSW729E, KRas, SOScat, and SOSW729E were separately buffer exchanged into 100 mM ammonium acetate (pH 7.4) using a Micro BioSpin 6 column (Bio-Rad). The GTP- or GDP-loaded KRas was mixed with SOScat or SOSW729E at 3:1 molar ratio in ammonium acetate and immediately analyzed using native MS. For BODIPY-GTP and Mant-GTPS experiments, 2 and 5 µM of the fluorophore analogs were added to the preformed mixture of SOScat and KRas–GTP, respectively (Fig. 1D), and were immediately analyzed using native MS. Additional details are available in SI Appendix, Materials and Methods for inhibitor binding assays.
Native MS.
Protein samples were introduced into an Exactive Plus with extended mass range (EMR) Orbitrap MS (Thermo Fisher Scientific). Samples were loaded into pulled borosilicate glass capillaries prepared in-house, and electrosprayed into the instrument with the voltage applied using a platinum wire directly inserted into the solution. Instrument parameters (SI Appendix, Table S3) were tuned to minimize gas phase activation and to preserve noncovalent interactions between KRas and SOScat or SOSW729E. All of the measurements were taken in triplicate and repeated on different days. The mole fractions of free SOScat, binary and ternary complexes were determined using Unidec software (50) for all triplicates, and average values were used for circle graphs. The additional details on CID, SID, and IM-MS experiments are found in SI Appendix, Materials and Methods.
Nucleotide Exchange Assay.
The rate of nucleotide dissociation was determined using KRas loaded with BODIPY-GDP in the absence and presence of SOScat using Clariostar BMG Labtech. The nucleotide exchange was initiated by adding excess amount of GTP ± KRas–GTP, KRasG13D–GTP, or KRasQ61H–GTP. The details on the assay can be found in SI Appendix, Materials and Methods.
Cryo-EM.
To obtain the ternary complex of KRasG13D and SOScat, first KRasG13D was loaded with GppNp (Guanosine 5′-[,-imido] triphosphate trisodium) and excess nucleotide was removed by HiTrap desalting (5 mL; GE) column and concentrated to 15 mg/mL The complex was formed by incubation of threefold molar excess of KRasG13D–GppNp with SOScat for 1 h in ice followed by size-exclusion chromatography to purify the complex to homogeneity. Additional details on data collection for single-particle cryo-EM, image processing, model building, and refinement are available in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. John Kuriyan for generously providing plasmids to express SOS fragments. This work was supported by National Cancer Institute (NCI) and National Institute of General Medical Sciences (NIGMS) (Grant R01GM121751), NIGMS (Grant DP2GM123486), and National Institute of Health (NIH) (Grant P41GM128577). Part of this work was performed at the National Center for CryoEM Access and Training and the Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High-Resolution Cryo-Electron Microscopy Program (Grant U24 GM129539) and by grants from the Simons Foundation (SF349247) and New York State Assembly.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022403118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Downward J., Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer 3, 11–22 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Colicelli J., Human RAS superfamily proteins and related GTPases. Sci. STKE 2004, RE13 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karnoub A. E., Weinberg R. A., Ras oncogenes: Split personalities. Nat. Rev. Mol. Cell Biol. 9, 517–531 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vigil D., Cherfils J., Rossman K. L., Der C. J., Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat. Rev. Cancer 10, 842–857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prior I. A., Lewis P. D., Mattos C., A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457–2467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox A. D., Fesik S. W., Kimmelman A. C., Luo J., Der C. J., Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 13, 828–851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernández-Medarde A., Santos E., Ras in cancer and developmental diseases. Genes Cancer 2, 344–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stephen A. G., Esposito D., Bagni R. K., McCormick F., Dragging ras back in the ring. Cancer Cell 25, 272–281 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Milburn M. V., et al., Molecular switch for signal transduction: Structural differences between active and inactive forms of protooncogenic ras proteins. Science 247, 939–945 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Vetter I. R., Wittinghofer A., The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Goitre L., Trapani E., Trabalzini L., Retta S. F., The Ras superfamily of small GTPases: The unlocked secrets. Methods Mol. Biol. 1120, 1–18 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Gureasko J., et al., Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol. 15, 452–461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rojas J. M., Oliva J. L., Santos E., Mammalian son of sevenless Guanine nucleotide exchange factors: Old concepts and new perspectives. Genes Cancer 2, 298–305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Margarit S. M., et al., Structural evidence for feedback activation by Ras⋅GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112, 685–695 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Sondermann H., et al., Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell 119, 393–405 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Vo U., et al., Monitoring Ras interactions with the nucleotide exchange factor son of sevenless (Sos) using site-specific NMR reporter signals and intrinsic fluorescence. J. Biol. Chem. 291, 1703–1718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith M. J., Neel B. G., Ikura M., NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc. Natl. Acad. Sci. U.S.A. 110, 4574–4579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freedman T. S., et al., Differences in flexibility underlie functional differences in the Ras activators son of sevenless and Ras guanine nucleotide releasing factor 1. Structure 17, 41–53 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iversen L., et al., Molecular kinetics. Ras activation by SOS: Allosteric regulation by altered fluctuation dynamics. Science 345, 50–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostrem J. M., Shokat K. M., Direct small-molecule inhibitors of KRAS: From structural insights to mechanism-based design. Nat. Rev. Drug Discov. 15, 771–785 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Lu S., Jang H., Zhang J., Nussinov R., Inhibitors of Ras-SOS interactions. ChemMedChem 11, 814–821 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Hillig R. C., et al., Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction. Proc. Natl. Acad. Sci. U.S.A. 116, 2551–2560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann M. H., et al., BI-3406, a potent and selective SOS1-KRAS interaction inhibitor, is effective in KRAS-driven cancers through combined MEK inhibition. Cancer Discov. 11, 142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hilton G. R., Benesch J. L., Two decades of studying non-covalent biomolecular assemblies by means of electrospray ionization mass spectrometry. J. R. Soc. Interface 9, 801–816 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanucara F., Holman S. W., Gray C. J., Eyers C. E., The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat. Chem. 6, 281–294 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Allison T. M., Bechara C., Structural mass spectrometry comes of age: New insight into protein structure, function and interactions. Biochem. Soc. Trans. 47, 317–327 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Leney A. C., Heck A. J. R., Native mass spectrometry: What is in the name? J. Am. Soc. Mass Spectrom. 28, 5–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quintyn R. S., Yan J., Wysocki V. H., Surface-induced dissociation of homotetramers with D2 Symmetry yields their assembly pathways and characterizes the effect of ligand binding. Chem. Biol. 22, 583–592 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Gault J., et al., High-resolution mass spectrometry of small molecules bound to membrane proteins. Nat. Methods 13, 333–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daneshfar R., Kitova E. N., Klassen J. S., Determination of protein-ligand association thermochemistry using variable-temperature nanoelectrospray mass spectrometry. J. Am. Chem. Soc. 126, 4786–4787 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Cong X., et al., Determining membrane protein-lipid binding thermodynamics using native mass spectrometry. J. Am. Chem. Soc. 138, 4346–4349 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Benesch J. L., Sobott F., Robinson C. V., Thermal dissociation of multimeric protein complexes by using nanoelectrospray mass spectrometry. Anal. Chem. 75, 2208–2214 (2003). [DOI] [PubMed] [Google Scholar]
- 33.El-Baba T. J., et al., Melting proteins: Evidence for multiple stable structures upon thermal denaturation of native ubiquitin from ion mobility spectrometry-mass spectrometry measurements. J. Am. Chem. Soc. 139, 6306–6309 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Raab S. A., et al., Evidence for many unique solution structures for chymotrypsin inhibitor 2: A thermodynamic perspective derived from vT-ESI-IMS-MS measurements. J. Am. Chem. Soc. 142, 17372–17383 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cong X., Liu Y., Liu W., Liang X., Laganowsky A., Allosteric modulation of protein-protein interactions by individual lipid binding events. Nat. Commun. 8, 2203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moghadamchargari Z., et al., Intrinsic GTPase activity of K-RAS monitored by native mass spectrometry. Biochemistry 58, 3396–3405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu S., Jang H., Nussinov R., Zhang J., The structural basis of oncogenic mutations G12, G13 and Q61 in small GTPase K-Ras4B. Sci. Rep. 6, 21949 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall B. E., Yang S. S., Boriack-Sjodin P. A., Kuriyan J., Bar-Sagi D., Structure-based mutagenesis reveals distinct functions for Ras switch 1 and switch 2 in Sos-catalyzed guanine nucleotide exchange. J. Biol. Chem. 276, 27629–27637 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Hunter J. C., et al., Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol. Cancer Res. 13, 1325–1335 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Patgiri A., Yadav K. K., Arora P. S., Bar-Sagi D., An orthosteric inhibitor of the Ras-Sos interaction. Nat. Chem. Biol. 7, 585–587 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janes M. R., et al., Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Shima F., et al., In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc. Natl. Acad. Sci. U.S.A. 110, 8182–8187 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smock R. G., Gierasch L. M., Sending signals dynamically. Science 324, 198–203 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhett J. M., Khan I., O’Bryan J. P., Biology, pathology, and therapeutic targeting of RAS. Adv. Cancer Res. 148, 69–146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vatansever S., Erman B., Gümüş Z. H., Comparative effects of oncogenic mutations G12C, G12V, G13D, and Q61H on local conformations and dynamics of K-Ras. Comput. Struct. Biotechnol. J. 18, 1000–1011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li S., Balmain A., Counter C. M., A model for RAS mutation patterns in cancers: Finding the sweet spot. Nat. Rev. Cancer 18, 767–777 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Jeng H.-H., Taylor L. J., Bar-Sagi D., Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nat. Commun. 3, 1168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loree J. M., et al., Not all RAS mutations created equal: Functional and clinical characterization of 80 different KRAS and NRAS mutations. J. Clin. Oncol. 35, 3589 (2017). [Google Scholar]
- 49.Huang W. Y. C., et al., A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marty M. T., et al., Bayesian deconvolution of mass and ion mobility spectra: From binary interactions to polydisperse ensembles. Anal. Chem. 87, 4370–4376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson C. W., et al., Isoform-specific destabilization of the active site reveals a molecular mechanism of intrinsic activation of KRas G13D. Cell Rep. 28, 1538–1550.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.