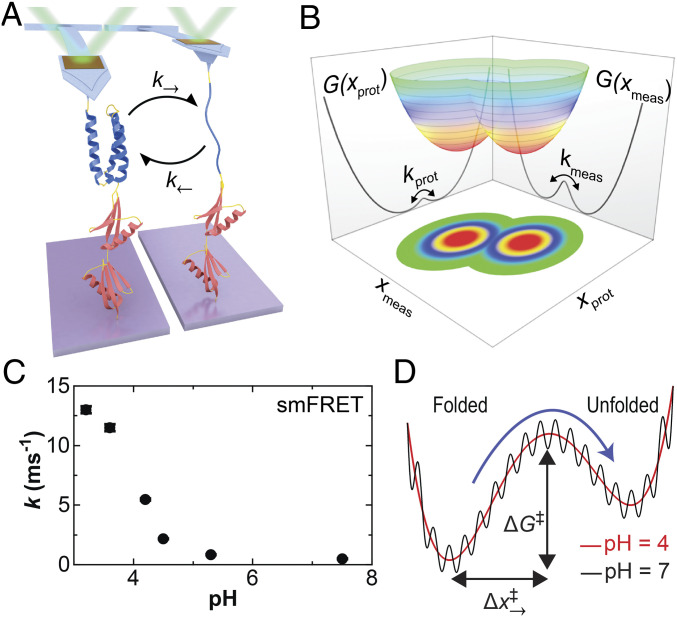
Fig. 1.
Probing the folding and unfolding dynamics of a globular protein by SMFS. (A) Cartoon showing a polyprotein consisting of a single copy of α3D (blue) and two copies of NuG2 (red) stretched with an atomic force microscope. At low forces, the mechanically labile α3D repeatedly unfolds and refolds as detected by a change in cantilever deflection. (B) A conceptual two-dimensional free-energy landscape shows the underlying protein extension (xprot) and the experimentally measured extension (xmeas). The macroscopic force probe has finite temporal resolution, and the application of force can introduce an entropic barrier between resolved states. (C) The sum of the equilibrium folding and unfolding rates for α3D in a strong denaturant (5 to 6 M urea) as a function of pH as determined in a prior smFRET study (37). (D) A conceptual sketch of α3D’s 1D free-energy landscape deduced by a combination of smFRET and molecular dynamics studies based on Ref. 37. The dramatic increase in α3D’s kinetics at low pH shown in panel C was explained as increased configurational diffusion along a smooth rather than a rough energy landscape.