Abstract
Simple Summary
Maedi-visna is a progressive wasting disease of sheep that leads to decreased animal condition and production. It is caused by Small Ruminant Lentiviruses (SRLV), single-stranded RNA viruses with high mutational potential. There is neither treatment nor vaccine against SRLV and proper diagnosis is the key element for efficient control measures against positive animals. This systematic review and meta-analysis summarizes the individual and flock prevalence of SRLV throughout the world and describes the diagnostic tests employed over the last four decades. Our results indicate that Europe is the continent with the most information on the prevalence of the infection as well as with the highest SRLV prevalence at the individual level. Flock prevalence depends directly on the individual prevalence. SRLV diagnostic methods in sheep have substantially changed during the last decades, but serologic methods have always been the most frequently used techniques for prevalence studies. A combination of at least two diagnostic tests is fully encouraged for future prevalence studies and health programs. ELISA and PCR show synergic effects in SRLV diagnosis.
Abstract
Small Ruminant Lentiviruses (SRLV) are highly prevalent retroviruses with significant genetic diversity and antigenic heterogeneity that cause a progressive wasting disease of sheep called Maedi-visna. This work provides a systematic review and meta-analysis of the last 40 years (1981–2020) of scientific publications on SRLV individual and flock prevalence. Fifty-eight publications and 314 studies were included. Most articles used a single diagnostic test to estimate prevalence (77.6%), whereas articles using three or more tests were scarce (6.9%). Serological tests are more frequently used than direct methods and ELISA has progressively replaced AGID over the last decades. SRLV infection in sheep is widespread across the world, with Europe showing the highest individual prevalence (40.9%) and being the geographical area in which most studies have been performed. Africa, Asia, and North America show values between 16.7% to 21.8% at the individual level. South and Central America show the lowest individual SRLV prevalence (1.7%). There was a strong positive correlation between individual and flock prevalence (ρ = 0.728; p ≤ 0.001). Despite the global importance of small ruminants, the coverage of knowledge on SRLV prevalence is patchy and inconsistent. There is a lack of a gold standard method and a defined sampling strategy among countries and continents.
Keywords: small ruminant lentiviruses, meta-analysis, prevalence, maedi-visna, seroprevalence
1. Introduction
Maedi-visna is a progressive wasting disease of sheep that causes important deleterious effects in animal production and limits animal trade worldwide [1,2,3]. This condition is caused by Small Ruminant Lentiviruses (SRLV), a group of single-stranded RNA viruses with high mutation and recombination potential [4]. Indeed, five main genotypes (A–E) and more than 28 subgroups have been already characterized [5]. This phylogenetic diversity implies high genetic and antigenic diversity, which hinder serologic and molecular diagnosis [6].
SRLV have tropism for the mononuclear-phagocyte system and induce slow, chronic, and persistent inflammation in four main target organs, namely lung, joints, nervous system, and mammary gland, inducing different clinical forms (i.e., pulmonary, articular, nervous, and mammary). Interestingly, the occurrence of each clinical form as well as the severity of the lesions depend on viral factors as well as the host immune response [1,3,7,8]. The most common issue after SRLV infection is increased replacement rates due to decreased animal condition and production [1].
There are no treatments or commercial vaccines for Maedi-visna. Thus, accurate diagnosis is the cornerstone for setting up an optimal control program of the infection and reduce its prevalence. Multiple diagnostic techniques can be used to detect SRLV infection. Indirect methods (AGID and ELISA) have been proposed as the most appropriate to detect infected animals, ELISA having higher sensitivity and lower specificity than AGID [9]. Direct methods to detect SRLV (PCR, indirect immunofluorescence, and in situ hybridization) are also efficient diagnostic techniques [10]. Recent studies have demonstrated the inherent inaccuracy of using a single diagnostic test [11], which is likely related to the wide SRLV antigenic diversity. However, host response can also play a role since animals from the same herd infected with the same SRLV exhibit significant differences in the susceptibility to infection and viral replication [12]. Furthermore, delay in seroconversion can be very variable among individuals [13]. Although initial descriptions of SRLV infection are from the 1950s [14], only during the last forty years has there been a growing body of publications assessing individual and flock SRLV prevalence around the world. However, a comprehensive compilation of the diagnostic methods used and their prevalence results in each continent is lacking.
The aim of this work is to estimate and compare the prevalence of SRLV in the world by performing a systematic review and meta-analysis of the articles published during the last 40 years (1981–2020) complemented by comprehensive description of diagnostic test used.
2. Materials and Methods
2.1. Literature Search and Recording of Information
Literature of the last 40 years (1981–2020, both included) dealing with SRLV prevalence in sheep was collected following PRISMA guidelines [15,16]. A flow diagram describing the selection process of references is detailed in Appendix A. Different databases were checked including PubMed, WOS, and Scopus. Keywords included: maedi, maedi-visna, maedi/visna, small ruminant lentivirus, SRLV, and/or prevalence. Criteria for eligibility were: (i) detailed information on SRLV prevalence in countries or regions within countries; (ii) abstract written or translated in English; and (iii) publication between 1981 and 2020, inclusive. The reference year of each prevalence study was the date on which the study was performed. Exceptionally, for publications in which this date was not available, the date of article publication was used. Criteria for exclusion were: (i) total number of sampled animals not detailed; and (ii) studies focused on diseased sheep.
Data from publications included in this meta-analysis were extracted by a single researcher (M.A.) and confirmed by three different investigators (R.d.M., A.R.L., and E.P.). Data items systematically collected are detailed in Appendix B.
2.2. Analyses
Qualitative epidemiological variables obtained from publications (i.e., presence or absence of data on animal prevalence, flock prevalence, and population size) and information on diagnostic techniques (i.e., presence or absence of data on sensitivity and specificity and number and type of tests used) were analyzed using contingency tables and represented as absolute and relative frequencies. Additionally, the type of diagnostic tests used for SRLV prevalence determinations was described over the four decades that this meta-analysis includes. A test was considered as diagnostic when applied to all samples collected or a randomly selected group of them. Graphs were produced with Prism 8.0.2 (GraphPad Software).
The five main statistical parameters used in this meta-analysis are detailed in Table 1. Apparent prevalence (percentage of positive animals) was used as most publications did not detail specificity and sensitivity of diagnostic tests, and consequently true prevalence (percentage of infected animals) could not be estimated. When the number of positive reactors to the test was not provided in the study, it was calculated with the prevalence and the sample size. When multiple diagnostic tests were performed, animals were considered positive if they were positive to at least one diagnostic test. Data obtained from publications were subdivided into two main groups: individual and flock prevalence. Moreover, prevalence data were grouped by continents and countries and values were calculated as the weighted arithmetic mean to attribute each study its relative importance, depending on sample size. The 95% confidence interval for the estimated prevalence values was calculated by using the formula of Wilson et al. [17]. This indicator provides a range of values in which the population prevalence can be found with a 95% degree of confidence. Heterogeneity of studies was quantified with the heterogeneity statistic I2. This parameter is based on Cochran’s Q test of homogeneity and provides useful information in the variability between the studies included in the meta-analysis. Additionally, an historical evolution of the infection by decades was performed. All the above-mentioned analyses were performed with Excel (Microsoft Office Professional Plus 2019, USA), except for heterogeneity statistic I2, which was calculated with OpenMeta[Analyst] software [18]. Maps were produced with GeoNames extension (DSAT, Microsoft Office Professional Plus 2019, United States) for Excel. Finally, correlation and determination coefficients between animal and flock prevalence were calculated with Spearman’s rank test by using IBM SPSS 19.0 for Windows (IBM Corporation, Armonk, NY, USA).
Table 1.
Definition of the parameters used in the epidemiological meta-analysis.
| Mathematical Equations |
|---|
| Apparent prevalence = (Pos/n)∙100 |
| Positive animals or flocks = (P∙n)/100 |
| Confidence interval = (2nP + Z_(α/2)^2 ± Z_(α/2) √(4nP(1 − P) + Z_(α/2)^2))/(2(n + Z(α/2)^2)) |
| Weighted arithmetic mean of prevalence = (∑ (Pi∙ni))/(∑ni) |
| Heterogeneity statistic I2 = ((Q − df)/Q) × 100 |
Pos, number of positive animals; n, sample size; P, prevalence value; Z value, varies depending on the percent of confidence; α, alpha level or difference between 100% and the confidence interval; Pi, prevalence value of each study to be averaged; ni, sample size of each study to be averaged; Q, Cochran’s homogeneity test statistic; df, degrees of freedom.
3. Results
3.1. Analysis of Publications and Diagnostic Tests
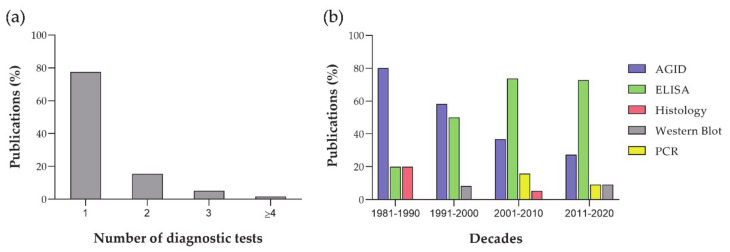
In total, 58 publications were included in this meta-analysis (Appendix A). All these publications showed individual prevalence studies, whereas 65.5% (38/58) also contained flock prevalence studies. In total, 314 prevalence studies were recorded. Information on the total number of animals and flocks in the studied geographical area (global population size) was only provided in 31% (18/58) and 34.2% (13/38) of publications, respectively. Sensitivity and specificity of the diagnostic tests were detailed in 36.2% (21/58) of publications. Most articles used a single diagnostic test (77.6%, 45/58) to calculate prevalence, whereas articles using three or more tests were scarce (6.9%, 4/58; Figure 1a). AGID was the most common diagnostic test from 1981 to 2000, showing a decreased importance over the years (Figure 1b). The use of ELISA showed a marked increase from 1981 to 2000, becoming the most important technique from 2001 to 2020, with constant values over the two decades. PCR has been used as a diagnostic tool for prevalence studies from 2001 to 2020, but there is a scarcity of publications using this technique as the main diagnostic tool. Histology and Western blot have been occasionally used as diagnostic tools.
Figure 1.
Analysis of diagnostic tests: (a) number of diagnostic tests performed in each article; and (b) evolution of the diagnostic tests from 1981 to 2020.
3.2. Individual SRLV Prevalence
Prevalence of SRLV infection in sheep per continent is provided in Table 2. Europe shows by far the highest value for SRLV prevalence (40.9%), whereas Africa, Asia, and North America show values in a range between 16.7% to 21.8%. South and Central America show the lowest individual SRLV prevalence (1.7%). Interestingly, results for Europe derive from 65 studies of SRLV prevalence in 22 publications with a total of 407,509 sheep tested during the last 40 years, whereas Africa only shows five studies in foru publications and a total of 1688 animals studied during the last four decades. All continents showed marked heterogeneity among studies, with prevalence values ranging from 0% to 71.2%. Appendix C provides an evolution of individual SRLV prevalence along decades in each continent.
Table 2.
Individual SRLV prevalence in each continent. Data extracted from scientific literature published from 1981 to 2020.
| Continent | Studies | N 1 | Prevalence (%) | Range (%) | Heterogeneity | Refs | ||
|---|---|---|---|---|---|---|---|---|
| Mean | CI 95% 2 | Min | Max | I2 3 (p Value) | ||||
| Africa | 5 | 1688 | 16.65 | 14.95–18.50 | 1.37 | 24.80 | 98.09 (<0.001) | [19,20,21,22] |
| Asia | 47 | 8309 | 20.38 | 19.52–21.26 | 0.00 | 71.20 | 98.60 (<0.001) | [23,24,25,26,27,28,29,30,31,32,33,34,35] |
| Europe | 65 | 407,509 | 40.90 | 40.75–41.05 | 0.00 | 66.43 | 98.98 (<0.001) | [11,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56] |
| North America | 46 | 124,542 | 21.76 | 21.53–21.99 | 0.00 | 52.00 | 99.61 (<0.001) | [57,58,59,60,61,62,63,64,65,66] |
| South and Central America | 41 | 46,418 | 1.67 | 1.56–1.79 | 0.00 | 30.00 | 91.52 (<0.001) | [67,68,69,70,71,72,73,74,75] |
| Global | 204 | 588,466 | 33.39 | 33.27–33.51 | 0.00 | 71.20 | 99.95 (<0.001) | |
1 N, number of animals tested; 2 CI 95%, 95% confidence interval; 3 I2, heterogeneity statistic.
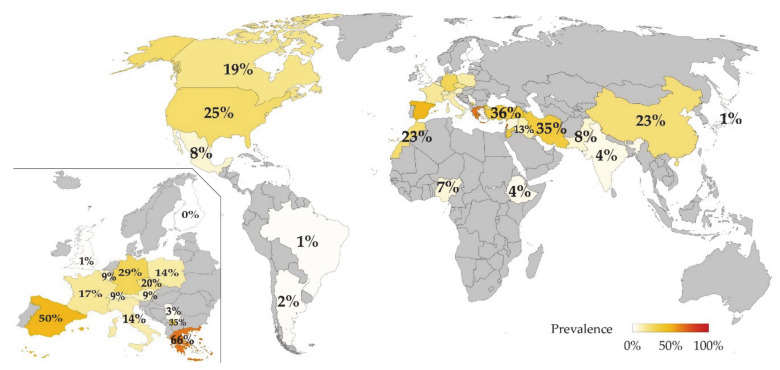
Infected animals per country are detailed in Figure 2 and extended information is provided in Appendix D. In total, 33 countries provided studies with valid data and were located in: Africa (n = 3), Asia (n = 10), Europe (n = 14), North America (n = 3), and South and Central America (n = 3). Countries with the highest individual SRLV prevalence are Lebanon, Greece, and Spain. Spain is the country with the highest number of animals studied (308,858).
Figure 2.
Individual SRLV prevalence (%) in sheep per country. Data extracted from scientific literature published from 1981 to 2020 and detailed in Table 2. Inset: Higher magnification of Europe.
3.3. Flock SRLV Prevalence
Prevalence of flock SRLV infection per continent is provided in Table 3. Asia is the continent showing the highest flock prevalence (66%), whereas Europe and North America are within a range of 44.4–48.6%. Africa is the continent showing a lesser percentage of prevalence (7.7%). Flock SRLV prevalence showed marked heterogeneity among studies in all continents. Appendix E provides an evolution of SRLV prevalence along decades in each continent. Distribution of flock SRLV prevalence along decades parallels the temporal evolution of individual prevalence.
Table 3.
Flock SRLV prevalence in each continent. Data extracted from scientific literature published from 1981 to 2020.
| Continent | Studies | N 1 | Prevalence (%) | Range (%) | Heterogeneity | Refs | ||
|---|---|---|---|---|---|---|---|---|
| Mean | CI 95% 2 | Min | Max | I2 3 (p Value) | ||||
| Africa | 1 | 13 | 7.69 | 1.37–33.31 | 7.69 | 7.69 | N/A 4 | [21] |
| Asia | 8 | 197 | 65.99 | 59.12–72.24 | 21.43 | 100.00 | 95.58 (<0.001) | 5 |
| Europe | 35 | 4590 | 44.38 | 42.95–45.82 | 0.00 | 100.00 | 99.98 (<0.001) | 6 |
| North America | 32 | 1933 | 48.58 | 46.35–50.81 | 0.00 | 100.00 | 95.26 (<0.001) | 7 |
| South and Central America | 34 | 2358 | 18.87 | 17.34–20.50 | 0.00 | 100.00 | 83.15 (<0.001) | 8 |
| Global | 110 | 9091 | 39.07 | 38.07. 40.08 | 0 | 100 | 99.94 (<0.001) | |
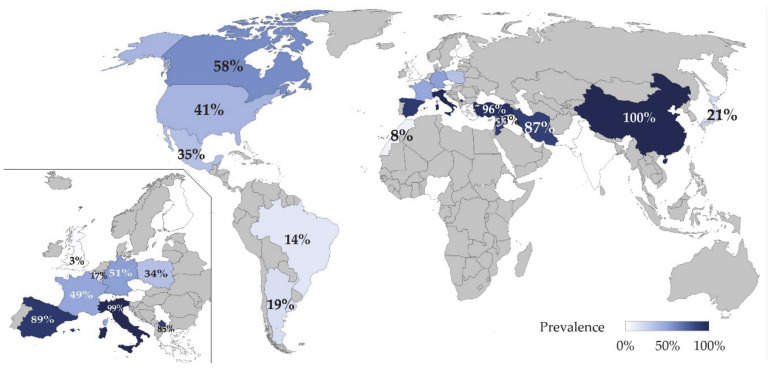
Infected flocks per country are detailed in Figure 3 and extended information is provided in Appendix F. In total, 23 countries provided studies with valid data and were located in: Africa (n = 1), Asia (n = 7), Europe (n = 9), North America (n = 3), and South and Central America (n = 3). Countries with the highest SRLV flock prevalence are Lebanon and China, whereas Poland is the country with the highest number of flocks studied (1621).
Figure 3.
Flock prevalence (%) of SRLV in sheep by country. Data extracted from scientific literature published from 1981 to 2020 and detailed in Table 3. Inset: Higher magnification of Europe.
3.4. Correlation between Individual and Flock SRLV Prevalence
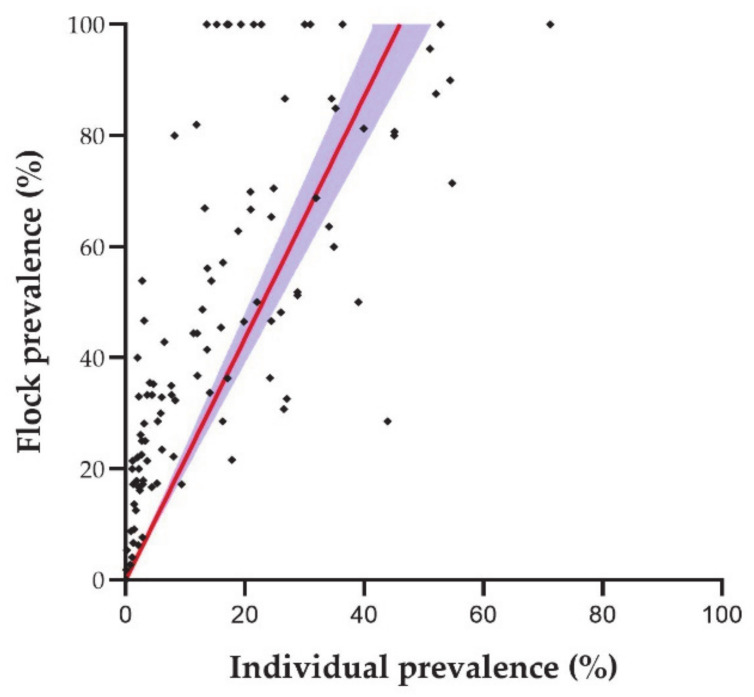
Individual and flock prevalence was obtained from 118 studies. There was a strong positive correlation between individual and flock prevalence (ρ = 0.728; p ≤ 0.001). Linear regression (y = 2.174x) demonstrated that each increase in individual prevalence induced at least two-fold increase in flock prevalence. Indeed, in some cases, flock prevalence reached 60% when individual prevalence was below 30% (Figure 4). The determination coefficient was 0.530.
Figure 4.
Correlation between individual and flock SRLV prevalence: dots are the data of each individual study (n = 118); the red line is the linear regression; and the blue area is the 95% confidence interval of the linear regression [11,21,25,26,28,29,31,33,35,36,37,38,41,42,45,47,48,49,51,52,54,57,58,59,61,62,63,64,65,66,67,69,71,72,73,74,75].
4. Discussion
This meta-analysis based on published research presents the distribution and prevalence of SRLV considering individual animals and flocks in the world over the last 40 years. The results indicate a widespread SRLV infection in all continents and underline the importance of SRLV in sheep throughout the world. SRLV is heterogeneously distributed, with marked variations not only between continents but also between regions in the same continent. This heterogeneity between studies reflects the multiple factor that influence SRLV prevalence.
Europe is the continent with most information on prevalence and distribution of infection as 1/3 of publications included in the study and 2/3 of animals analyzed belong to this continent. Recent phylogenetic studies suggest that SRLV-genotype A, historically associated with Maedi-visna in sheep, may have arisen in a territory within the current borders of Turkey and spread across the world with human migratory movements [25,76]. First reports of lesions compatible with those caused by SRLV were likely reported in The Netherlands [77] and the description of the disease together with the infection took place in Iceland [78]. This could explain the marked interest of European countries in the study of this infection. North America and Asia also show a notable SRLV prevalence and a growing number of published studies in both continents. There are striking differences among countries in the number of animals tested against SRLV, Spain being the country with highest number of studied animals (n = 308,858) and Pakistan the country with the fewest tested sheep (n = 93). Interpretation of prevalence studies with low sample size should be performed cautiously.
SRLV prevalence data depend on the different routes for viral spread. Horizontal (aerosols and direct contact) transmission is the main route for SRLV propagation, and it is likely the route responsible for most of the SRLV clinical cases [1]. This route is influenced by numerous environmental, demographic, and management factors [1,65]. Additionally, vertical lactogenic transmission also plays an important epidemiological role, with up to 16% of lambs born from seropositive ewes being infected during their first day of life [79]. The high individual SRLV prevalence found in Europe is likely associated with high density of ovine populations and intensive management. For instance, Greece and Spain show the highest prevalence values in Europe and both countries are within the top ten countries in milk production [80], which is usually performed in intensive management systems and implies continuous close contact within animals. However, Italy is also among this top ten countries, but its prevalence is not that high. Average flock sizes in dairy sheep are similar in Greece, Spain, and Italy (140–161 ewes/farm) [81], thus this factor cannot explain the Italian moderate SRLV prevalence values. Note that Europe is the third continent in the ranking of flock prevalence despite being the first in individual prevalence. This decrease might be associated to the several control and eradication programs performed in European flocks during the last decades.
Iran, Turkey, China, Jordan, and Lebanon are the Asiatic countries with the highest SRLV individual prevalence. Indeed, Iran, Turkey, and China are major producers of meat and milk [80,82], further highlighting the relevance of the production type and management system [83,84,85]. Based on the FAO database, Asia is the continent with the highest ovine population of the world and this fact could be determinant in SRLV transmission between flocks, as Asia is the continent with the highest flock SRLV prevalence. Interestingly, most ovine breeding stocks are located in China and India and individual SRLV prevalence in these countries is strikingly different, pointing out that the total number of animals in a given geographical area is not a relevant factor for intra-flock SRLV transmission. Differences between individual prevalence values of these two countries could be likely explained by differences in management systems, being mainly semi-intensive to intensive in the former and extensive to nomadic in the latter [80,86]. A similar explanation could be applied to the differences found between individual prevalence values in African countries, where Morocco shows much higher values than Ethiopia and Nigeria, for instance. About 75% of sheep in Ethiopia are kept on small-scale mixed farms, with an average number of 5.3 sheep per farm, usually raised in privately owned land [80,87,88]. The Animal and Plant Health Inspection Service of the United States Department of Agriculture provides an outstanding explanation on the influence of cultural, geographical and management factors in SRLV prevalence in North America [65] showing the significant role of transport (i.e., crowding of animals) and the knowledge and concern of farmers about the disease. Finally, low values of individual SRLV prevalence in South America are likely associated with the low number of animals in this region together with management factors, mostly extensive rearing [83,84,85].
The results of this meta-analysis greatly depend on the generation of prevalence data and their publication in scientific repositories. Indeed, a discrepancy between OIE reports and the information available in the scientific literature on SRLV prevalence was noted. Based on the information provided by the OIE (Disease Timelines of the World Animal Health Information Database, WAHIS interface [17]), there are 28 additional countries (Andorra, Bosnia and Herzegovina, Bulgaria, Chile, Colombia, Comoros, Croatia, Cyprus, Denmark, Estonia, North Macedonia, Hungary, Israel, Latvia, Liechtenstein, Luxembourg, Malta, Mexico, Mongolia, Netherlands, Norway, Autonomous Palestinian Territories, Portugal, Romania, Russia, Slovakia, Slovenia, and Sweden.) with SRLV infection in sheep that are absent of our study because no public publications from those countries were found, or they did not fulfilled the inclusion criteria. On the contrary, from 2005 to 2019, China, Costa Rica, Iran, Morocco, and Pakistan have never reported SRLV infection to the OIE and India, Lebanon, Czech Republic, Syria, and Turkey appear as “disease absent” despite available scientific descriptions from all these countries [20,21,23,24,25,26,27,29,30,33,34,35,55,67]. First reports on SRLV infection in Brazil according to OIE are from 2017, whereas scientific publications already reported the disease in 2007 [69]. Therefore, publicly available scientific literature might not reflect the real situation in different countries; for instance, a prevalence of 0.7% is the only datum available from the UK [42], but the infection seems to be much more widespread, reaching a high number of flocks and individual animals. Indeed, recent studies in Scottish flocks with a novel multiplex ELISA (MVD-Enferplex GSMD multiplex ELISA Kit, MV Diagnostics, Edinburgh, UK) revealed an individual prevalence of 11.7% (sample size: 2659 animals) and SRLV infection present in 15 out of 17 studied flocks (N. Watt, unpublished data, 2020). Accordingly, flocks with more than 10 years within the Scottish maedi-visna control scheme showed spontaneous outbreaks of disease with up to 90% of animals infected [89,90].
As expected, the results clearly indicate that flock prevalence is linked to individual prevalence. Indeed, flock prevalence generally doubles individual prevalence. Interestingly, some infected areas show a 100% prevalence in flocks while the infection is just about 20% in animals, suggesting that the multiplying factor could be higher than two-fold under certain conditions. Increased prevalence is related to any activity that may imply prolonged close contact between animals favoring horizontal transmission such as intensive production system [1,65], transportation, or sharing milking machines [43,91]. Flock size might also play a role as higher numbers of animals per flock relate to higher prevalence [59,61].
SRLV diagnostic methods in sheep have substantially changed during the last decades but serologic methods have always been the most used techniques in prevalence studies. AGID was the most common technique in the 1980s and 1990 as it was the recommended test by the OIE for regulatory purposes [9]. However, from the beginning of the 21st century, several ELISA tests have replaced AGID as the most reliable method. Interestingly, most of the publications included in this meta-analysis used only a single diagnostic test to estimate the SRLV prevalence. The use of a single test has proved to underestimate the number of infected animals, impairing proper segregation of infected and non-infected individuals, which leads to a slower control of the infection and hinders accurate analysis of productive and clinicopathological parameters [11,25]. In the current situation of uncertainty regarding circulating SRLV strains, the most reliable/efficient strategy to identify infected animals would involve performing at least two diagnostic tests in parallel. The election of the most suitable diagnostic tests should be carefully considered based on the most prevalent circulating strain/s in a geographical area. The first test should be ELISA-based as they are highly sensitive and specific, thus suitable for high-throughput testing [9]. However, sensitivity and specificity of any ELISA diagnostic test should not be considered universal due to the scarcity of cross-reacting antibodies among different SRLV genotypes [92,93]. Serologic approaches may have disadvantages as they cannot detect animals with low antibody titers, that can remain as carriers and potentially cause disease outbreaks [94]. The second test should be complementary to the first one and targeted towards specific animal populations. For instance, most of the publications analyzed here excluded animals younger than six months as colostral antibodies can interfere with serologic testing. This could imply overlooking an important group of animals that are pivotal for SRLV transmission [79,95]. Direct methods such as PCR could mitigate ELISA drawbacks by detecting the viral load peak found in infected lambs during the neonatal period [62,96]. Therefore, using a direct technique as a second diagnostic test can help to detect recently-infected animals without a detectable serologic response [25,97]. A combination of ELISA and PCR has already been proposed as a synergic way to accurately diagnose SRLV infection, as it provides excellent results and improves the accuracy of the diagnosis in both, acute and chronic infections [13,22,31,32,98]. Until recently, a commercial diagnostic PCR was not available commercially [11] despite several publications setting up different protocols [99]. Therefore, this strategy can increase diagnosis sensitivity and potentially imply success in SRLV control strategies. However, it could simultaneously increase costs and, potentially, reduce diagnosis specificity. In any case, this strategy has proven to be useful as it has demonstrated the infection in flocks that were previously declared as uninfected and it has improved the sensitivity of the diagnosis in countries such as Spain [11,100], UK [89], and Switzerland [101].
This analysis is based on multiple publications with important differences in the study design and efforts to group publications based on animal information (breed, sex, or age) were fruitless. Despite this heterogeneity between SRLV prevalence, the present meta-analysis provides a unique and valuable approach to worldwide SRLV distribution. Moreover, review of individual publications can help to dissect the influence of these individual parameters. For instance, breed is clearly a predisposing factor in multiple geographic areas [19,25,59]. In this sense, genetic selection of resistant animals has been proposed as a control method, with TMEM154 as a promising target gene [102]. However, a recent study demonstrates that control measures based on a single gene may not be as useful as expected [100]. Age also seems to be related to higher prevalence of infection, with a non-linear increase that reaches the maximum at about four years of age [59,61,65]. Influence of sex in prevalence is not that obvious, some studies indicate males being more predisposed than females [43], whereas others indicate non-significant differences [57,59,63].
Limitations of this work arise from the diversity of study designs and data expression. Specificity and sensitivity of the test was only specified in 36.2% of publications and we decided to deal with apparent prevalence to avoid disregarding most of the selected publications. Technical information of the diagnostic test employed should be stated whenever possible to ease further data analysis [103,104]. Further studies investigating the sensitivities and specificities of the test that were not provided in the studied publications will be useful and interesting so that further analysis of these data based on real prevalence values could be performed. Unfortunately, recurring prevalence studies in the same geographical area are scarce. A publication bias might exist when SRLV infection is discovered in a certain area, likely leading to an increased number of reports, paralleling the increase of scientific interest. In areas where the disease is enzootic, the interest might not be similar.
5. Conclusions
SRLV infection in sheep is widespread across the world, Europe showing the highest individual prevalence and being the geographical area in which more studies have been performed. Prevalence of infected flocks shows a strong correlation with the individual prevalence. Most studies are based on a single diagnostic test, implying a risk of underestimating the real infection prevalence. Serological tests are more commonly used than direct methods and, among them, ELISA has progressively replaced AGID along the last two decades. There is a moderate disagreement between the information reported to the OIE and the scientific literature. This review highlights the need for more systematic and frequent prevalence studies using a consistent testing strategy.
Acknowledgments
Julia Barrio is fully acknowledged for her wise advice in data acquisition.
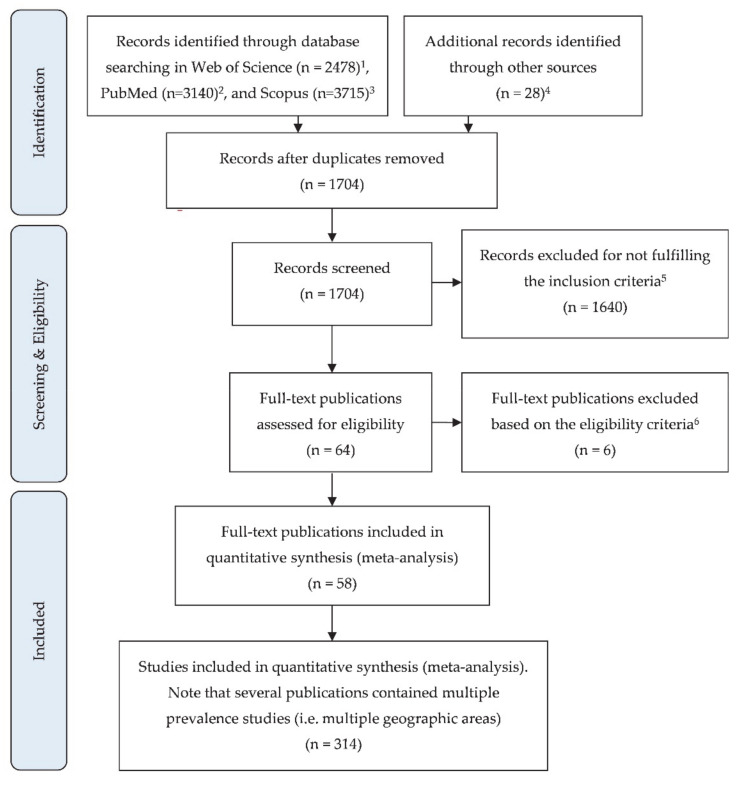
Appendix A
Flow diagram indicating procedure for identification, screening, selection and inclusion of publications in this meta-analysis. Modified from the PRISMA Statement [93].
Figure A1.
1 Keywords in Web of Science. maedi: 1641; maedi prevalence: 196; maedi-visna: 161; maedi-visna prevalence: 161; maedi/visna: 161; SRLV: 40; SRLV prevalence: 40; small ruminant lentivirus: 39; small ruminant lentivirus prevalence: 39. 2 Keywords in Pubmed. maedi: 733; maedi prevalence: 156; maedi-visna: 709; maedi-visna prevalence: 152; maedi/visna: 709; SRLV: 189; SRLV prevalence: 73; small ruminant lentivirus: 328; small ruminant lentivirus prevalence: 91. 3 Keywords in Scopus. maedi: 1019; maedi prevalence: 189; maedi-visna: 578; maedi-visna prevalence: 150; maedi/visna: 968; SRLV: 281; SRLV prevalence: 85; small ruminant lentivirus: 326; small ruminant lentivirus prevalence: 119. 4 Other sources. Conference Proceedings: 9; Google Scholar: 19. 5 Eligibility criteria: Information on SRLV prevalence; abstract available in English; publication from 1981 to 2020, inclusive. 6 Exclusion criteria: Total number of sampled animals not detailed; study focused on diseased sheep.
Appendix B
The following is the information that was systematically extracted from the publications included in the meta-analysis.
-
Information related to the publication time and location:
-
◦
Country of the study
-
◦
Continent of the study
-
◦
Year of the study
-
◦
Year of the publication
-
◦
-
Information related to diagnostic procedures:
-
◦
Number of techniques employed in the study
-
◦
Type of diagnostic technique
-
◦
Sensitivity and specificity of tests
-
◦
-
Information related to epidemiological data of the population:
-
◦
Total number of animals in the geographical area of the study
-
◦
Total number of flocks in the geographical area of the study
-
◦
-
Information related to epidemiological data of the sample:
-
◦
Sample size
-
◦
Number of positive animals and/or flocks (A flock was considered positive when one or more animals were positive to SRLV test)
-
◦
Subgroups of samples within the same publication. (i.e., prevalence in different regions, which were considered as independent studies)
-
◦
-
Information related to production system factors:
-
◦
Flock size
-
◦
Type of management system
-
◦
-
Information related to the animal factors:
-
◦
Breed of sampled animals
-
◦
Sex of sampled animals
-
◦
Age of sampled animals
-
◦
Appendix C
Individual SRLV prevalence in each continent by decades. Mean, number of animals tested (N), 95% Confidence Interval (CI 95%), and range are provided.
Table A1.
Animal SLRV prevalence in each continent from 1981 to 1990.
| Continent | Studies | N | Prevalence | Range | ||
|---|---|---|---|---|---|---|
| Mean | CI 95% | Min | Max | |||
| Africa | 1 | 72 | 2.78 | 0.77–9.57 | 2.78 | 2.78 |
| Asia | 0 | 0 | N/A 1 | N/A | N/A | N/A |
| Europe | 5 | 24,969 | 16.34 | 15.88–16.80 | 1.70 | 26.67 |
| North America | 19 | 57,593 | 19.13 | 18.81–19.45 | 0.00 | 39.92 |
| South and Central America | 0 | 0 | ||||
| Global | 25 | 82,634 | 18.27 | 18.01–18.54 | 0.00 | 39.92 |
1 N/A, not applicable.
Table A2.
Animal SLRV prevalence in each continent from 1991 to 2000.
| Continent | Studies | N | Prevalence | Range | ||
|---|---|---|---|---|---|---|
| Mean | CI 95% | Min | Max | |||
| Africa | 2 | 1267 | 20.99 | 18.84–23.32 | 6.74 | 24.80 |
| Asia | 13 | 2890 | 6.02 | 5.21–6.95 | 0.00 | 12.20 |
| Europe | 11 | 26,232 | 9.10 | 8.76–9.45 | 0.00 | 24.00 |
| North America | 21 | 39,444 | 26.49 | 26.05–26.92 | 2.19 | 52.00 |
| South and Central America | 0 | 0 | N/A 1 | N/A | N/A | N/A |
| Global | 47 | 69,833 | 19.01 | 18.72–19.30 | 0.00 | 52.00 |
1 N/A, not applicable.
Table A3.
Animal SLRV prevalence in each continent from 2001 to 2010.
| Continent | Studies | N | Prevalence | Range | ||
|---|---|---|---|---|---|---|
| Mean | CI 95% | Min | Max | |||
| Africa | 2 | 349 | 3.72 | 2.19–6.27 | 1.37 | 5.42 |
| Asia | 17 | 3889 | 28.57 | 27.17–30.01 | 0.00 | 68.97 |
| Europe | 5 | 296,695 | 51.25 | 51.07–51.43 | 24.80 | 66.43 |
| North America | 3 | 23,923 | 22.21 | 21.69–22.74 | 1.95 | 28.82 |
| South and Central America | 18 | 26,529 | 1.09 | 0.97–1.22 | 0.00 | 4.42 |
| Global | 45 | 351,385 | 45.19 | 45.02–45.35 | 0.00 | 68.97 |
Table A4.
Animal SLRV prevalence in each continent from 2011 to 2020.
| Continent | Studies | N | Prevalence | Range | ||
|---|---|---|---|---|---|---|
| Mean | CI 95% | Min | Max | |||
| Africa | 0 | 0 | N/A 1 | N/A | N/A | N/A |
| Asia | 17 | 1530 | 26.67 | 24.51–28.94 | 4.29 | 71.20 |
| Europe | 44 | 59,613 | 13.65 | 13.38–13.93 | 0.00 | 54.73 |
| North America | 3 | 3582 | 8.93 | 8.04–9.91 | 4.63 | 15.19 |
| South and Central America | 23 | 19,889 | 2.45 | 2.24–2.67 | 0.00 | 30.00 |
| Global | 87 | 84,614 | 11.05 | 10.84–11.27 | 0.00 | 71.20 |
1 N/A, not applicable.
Appendix D
Table A5.
Individual SRLV prevalence in each country. Mean, 95% confidence interval, and range are provided. Data extracted from scientific literature published from 1981 to 2020.
| Country | Studies | N 1 | Prevalence (%) | Range (%) | Refs | ||
|---|---|---|---|---|---|---|---|
| Mean | CI 95% 2 | Min | Max | ||||
| AFRICA | |||||||
| Ethiopia | 2 | 349 | 3.72 | 2.19–6.27 | 1.37 | 5.42 | [19] |
| Morocco | 2 | 1072 | 23.32 | 20.89–25.94 | 2.78 | 24.80 | [20,21] |
| Nigeria | 1 | 267 | 6.74 | 4.31–10.40 | 6.74 | 6.74 | [22] |
| ASIA | |||||||
| China | 12 | 672 | 22.77 | 19.76–26.09 | 5.36 | 50.00 | [29] |
| India | 1 | 140 | 4.29 | 1.98–9.03 | 4.29 | 4.29 | [24] |
| Iraq | 1 | 210 | 12.86 | 8.99–18.06 | 12.86 | 12.86 | [32] |
| Iran | 1 | 220 | 34.55 | 28.58–41.05 | 34.55 | 34.55 | [26] |
| Japan | 4 | 771 | 0.78 | 0.36–1.69 | 0.00 | 1.24 | [31] |
| Jordan | 1 | 231 | 36.36 | 30.43–42.74 | 36.36 | 36.36 | [28] |
| Lebanon | 1 | 184 | 71.20 | 64.27–77.25 | 71.20 | 71.20 | [33] |
| Pakistan | 1 | 93 | 7.53 | 3.69–14.73 | 7.53 | 7.53 | [34] |
| Syria | 13 | 2890 | 6.02 | 5.21–6.95 | 0.00 | 12.20 | [35] |
| Turkey | 12 | 2898 | 35.51 | 33.79–37.27 | 0.00 | 68.97 | [23,25,27,30] |
| EUROPE | |||||||
| Austria | 1 | 883 | 9.40 | 7.65–11.50 | 9.40 | 9.40 | [56] |
| Belgium | 1 | 555 | 9.37 | 7.22–12.08 | 9.37 | 9.37 | [54] |
| Czech Republic | 1 | 2801 | 19.85 | 18.41–21.37 | 19.85 | 19.85 | [55] |
| Finland | 1 | 10,802 | 0.00 | 0.00–0.04 | 0.00 | 0.00 | [45] |
| France | 3 | 23,404 | 16.68 | 16.20–17.16 | 1.70 | 26.67 | [41] |
| Germany | 2 | 2252 | 28.51 | 26.68–30.41 | 0.00 | 28.80 | [46,51] |
| Greece | 1 | 143 | 66.43 | 58.35–73.65 | 66.43 | 66.43 | [53] |
| Italy | 1 | 682 | 13.64 | 11.26–16.42 | 13.64 | 13.64 | [49] |
| Kosovo | 6 | 10,544 | 35.19 | 34.28–36.10 | 12.92 | 45.93 | [48] |
| Poland | 16 | 19,253 | 14.33 | 13.84–14.83 | 0.16 | 54.73 | [39,52] |
| Serbia | 14 | 11,709 | 3.38 | 3.07–3.73 | 0.00 | 9.96 | [43] |
| Spain | 16 | 308,858 | 49.84 | 49.67–50.02 | 1.23 | 54.41 | [11,36,37,38,40,47,50] |
| Switzerland | 1 | 3866 | 9.00 | 8.14–9.94 | 9.00 | 9.00 | [44] |
| United Kingdom | 1 | 11,757 | 0.74 | 0.60–0.91 | 0.74 | 0.74 | [42] |
| NORTH AMERICA | |||||||
| Canada | 29 | 68,019 | 19.21 | 18.92–19.51 | 0.00 | 50.00 | [57,60,62,63,66] |
| Mexico | 1 | 157 | 7.64 | 4.43–12.88 | 7.64 | 7.64 | [64] |
| United States | 16 | 56,366 | 24.87 | 24.51–25.23 | 2.19 | 52.00 | [58,59,61,65] |
| SOUTH AND CENTRAL AMERICA | |||||||
| Argentina | 29 | 42,597 | 1.69 | 1.57–1.82 | 0.00 | 30.00 | [72,73] |
| Brazil | 6 | 3103 | 1.32 | 0.98–1.79 | 0.11 | 8.20 | [68,69,70,71,74,75] |
| Cosa Rica | 6 | 718 | 1.95 | 1.16–3.25 | 0.00 | 4.26 | [67] |
1 N, number of animals tested. 2 CI 95%, 95% confidence interval.
Appendix E
Flock SRLV prevalence in each continent by decades. Mean, number of flocks tested (N), 95% Confidence Interval (CI 95%), and range are provided.
Table A6.
Flock SLRV prevalence in each continent from 1981 to 1990.
| Continent | Studies | N | Prevalence | Range | ||
|---|---|---|---|---|---|---|
| Mean | CI 95% | Min | Max | |||
| Africa | 1 | 13 | 7.69 | 1.37–33.31 | 7.69 | 7.69 |
| Asia | 0 | 0 | N/A 1 | N/A | N/A | N/A |
| Europe | 4 | 477 | 58.49 | 54.02–62.83 | 12.50 | 98.94 |
| North America | 12 | 675 | 62.96 | 59.26–66.52 | 0.00 | 100.00 |
| South and Central America | 0 | 0 | N/A | N/A | N/A | N/A |
| Global | 17 | 1165 | 60.52 | 57.68–63.28 | 0 | 100 |
1 N/A, not applicable.
Table A7.
Flock SLRV prevalence in each continent from 1991 to 2000.
| Continent | Studies | N | Prevalence | Range | ||
|---|---|---|---|---|---|---|
| Mean | CI 95% | Min | Max | |||
| Africa | 0 | 0 | N/A 1 | N/A | N/A | N/A |
| Asia | 1 | 73 | 32.88 | 23.19–44.27 | 32.88 | 32.88 |
| Europe | 8 | 445 | 11.01 | 8.43–14.26 | 0.00 | 100.00 |
| North America | 15 | 357 | 53.22 | 48.04–58.34 | 20.00 | 100.00 |
| South and Central America | 0 | 0 | N/A | N/A | N/A | N/A |
| Global | 24 | 875 | 30.06 | 27.11–33.18 | 0.00 | 100.00 |
1 N/A, not applicable.
Table A8.
Flock SLRV prevalence in each continent from 2001 to 2010.
| Continent | Studies | N | Prevalence | Range | ||
|---|---|---|---|---|---|---|
| Mean | CI 95% | Min | Max | |||
| Africa | 0 | 0 | N/A 1 | N/A | N/A | N/A |
| Asia | 3 | 67 | 76.12 | 64.67–84.73 | 21.43 | 95.65 |
| Europe | 4 | 912 | 92.65 | 90.78–94.17 | 51.22 | 100.00 |
| North America | 2 | 759 | 34.91 | 31.61–38.37 | 22.08 | 36.36 |
| South and Central America | 17 | 1234 | 12.97 | 11.21–14.96 | 0.00 | 25.00 |
| Global | 26 | 2972 | 44.82 | 42.67–46.24 | 0.00 | 100.00 |
1 N/A, not applicable.
Table A9.
Flock SLRV prevalence in each continent from 2011 to 2020.
| Continent | Studies | N | Prevalence | Range | ||
|---|---|---|---|---|---|---|
| Mean | CI 95% | Min | Max | |||
| Africa | 0 | 0 | N/A 1 | N/A | N/A | N/A |
| Asia | 4 | 57 | 96.49 | 88.08–99.03 | 83.33 | 100.00 |
| Europe | 19 | 2756 | 31.35 | 29.64–33.11 | 2.75 | 100.00 |
| North America | 3 | 142 | 41.55 | 33.77–49.77 | 35.00 | 51.85 |
| South and Central America | 17 | 1124 | 25.36 | 22.90–27.98 | 0.00 | 100.00 |
| Global | 43 | 4079 | 30.96 | 29.56–32.40 | 0.00 | 100.00 |
1 N/A, not applicable.
Appendix F
Table A10.
Flock SRLV prevalence in each country. Mean, 95% confidence interval, and range are provided. Data extracted from scientific literature published from 1981 to 2020.
| Country | Studies | N 1 | Prevalence (%) | Range (%) | Refs | ||
|---|---|---|---|---|---|---|---|
| Mean | CI 95% 2 | Min | Max | ||||
| AFRICA | |||||||
| Ethiopia | 0 | 0 | N/A 3 | N/A | N/A | N/A | N/A |
| Morocco | 1 | 13 | 7.69 | 1.37–33.31 | 7.69 | 7.69 | [21] |
| Nigeria | 0 | 0 | N/A | N/A | N/A | N/A | N/A |
| ASIA | |||||||
| China | 1 | 24 | 100.00 | 86.20–100.00 | 100.00 | 100.00 | [29] |
| India | 0 | 0 | N/A | N/A | N/A | N/A | N/A |
| Iraq | 0 | 0 | N/A | N/A | N/A | N/A | N/A |
| Iran | 1 | 30 | 86.67 | 70.32–94.69 | 86.67 | 86.67 | [26] |
| Japan | 1 | 14 | 21.43 | 7.57–47.59 | 21.43 | 21.43 | [31] |
| Jordan | 2 | 17 | 88.24 | 65.66–96.71 | 83.33 | 100.00 | [28] |
| Lebanon | 1 | 16 | 100.00 | 80.64–100.00 | 100.00 | 100.00 | [33] |
| Pakistan | 0 | 0 | N/A | N/A | N/A | N/A | N/A |
| Syria | 1 | 73 | 32.88 | 23.19–44.27 | 32.88 | 32.88 | [35] |
| Turkey | 1 | 23 | 95.65 | 79.01–99.23 | 95.65 | 95.65 | [25] |
| EUROPE | |||||||
| Austria | 0 | 0 | N/A | N/A | N/A | N/A | N/A |
| Belgium | 1 | 87 | 17.24 | 10.74–26.52 | 17.24 | 17.24 | [54] |
| Czech Republic | 0 | 0 | N/A | N/A | N/A | N/A | N/A |
| Finland | 1 | 340 | 0.00 | 0.00–1.12 | 0.00 | 0.00 | [45] |
| France | 3 | 383 | 48.56 | 43.60–53.56 | 12.50 | 86.67 | [41] |
| Germany | 1 | 41 | 51.22 | 36.48–65.75 | 51.22 | 51.22 | [51] |
| Greece | 0 | 0 | N/A | N/A | N/A | N/A | N/A |
| Italy | 1 | 94 | 98.94 | 94.22–99.81 | 98.94 | 98.94 | [49] |
| Kosovo | 1 | 318 | 84.91 | 80.56–88.42 | 84.91 | 84.91 | [48] |
| Poland | 15 | 1621 | 34.24 | 31.97–36.58 | 3.77 | 71.43 | [39] |
| Serbia | 0 | 0 | N/A | N/A | N/A | N/A | N/A |
| Spain | 11 | 980 | 89.49 | 87.41–91.26 | 6.67 | 100.00 | [11,36,37,38,40,47] |
| Switzerland | 0 | 0 | N/A | N/A | N/A | N/A | N/A |
| United Kingdom | 1 | 726 | 2.75 | 1.79–4.22 | 2.75 | 2.75 | [79] |
| NORTH AMERICA | |||||||
| Canada | 15 | 849 | 58.30 | 54.96–61.58 | 0.00 | 100.00 | [57,62,63,66] |
| Mexico | 1 | 20 | 35.00 | 18.12–56.71 | 35.00 | 35.00 | [64] |
| United States | 16 | 1064 | 41.07 | 38.15–44.05 | 20.00 | 87.50 | [58,59,61,65] |
| SOUTH AND CENTRAL AMERICA | |||||||
| Argentina | 29 | 2239 | 18.94 | 17.37–20.61 | 0.00 | 100.00 | [72,73] |
| Brazil | 4 | 104 | 14.42 | 8.94–22.44 | 1.85 | 80.00 | [69,71,74,75] |
| Cosa Rica | 1 | 15 | 40.00 | 19.82–64.25 | 40.00 | 40.00 | [67] |
1 N, number of animals tested. 2 CI 95%, 95% confidence interval. 3 N/A, not applicable.
Author Contributions
Conceptualization M.A., M.P., R.R. (Ramsés Reina), I.d.B., and L.L.; data curation R.d.M., M.A., A.R.-L., I.E., R.R. (Raúl Resendiz), E.P., and H.R.; formal analysis R.d.M., M.A., A.R.-L., I.E., and I.d.B.; funding acquisition D.d.A., R.R. (Ramsés Reina), and L.L.; methodology R.d.M., M.A., R.R. (Ramsés Reina), I.d.B., and L.L.; supervision M.P., D.d.A., R.R. (Ramsés Reina), I.d.B., and L.L.; validation, I.d.B.; writing—original R.d.M., M.A., R.R. (Ramsés Reina), and L.L.; and writing—review and editing R.d.M., M.A., A.R.-L., I.E., R.R. (Raúl Resendiz), E.P., H.R., M.P., D.d.A., R.R. (Ramsés Reina), I.d.B., and L.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by grants from the Spanish Ministry of Science, Innovation and Universities (RTI2018-096172-B-C31 and RTI2018-096172-B-C33), FOGAIBA and the Recognized Research Groups of Government of Aragón (A17_20R, Animal Health and Reproduction). R.d.M. was a PhD student funded by the Department of Innovation, Research and University of Aragón. ARL and EP were PhD students funded by the Spanish Ministry of Education. IE was a PhD student funded by the Universidad Pública de Navarra. RResendiz was a PhD student funded by CONACYT.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the scientific publications listed in the Reference section.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Minguijón E., Reina R., Pérez M., Polledo L., Villoria M., Ramírez H., Leginagoikoa I., Badiola J.J., García-Marín J.F., de Andrés D., et al. Small ruminant lentivirus infections and diseases. Vet. Microbiol. 2015;181:75–89. doi: 10.1016/j.vetmic.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Juste R.A., Villoria M., Leginagoikoa I., Ugarte E., Minguijon E. Milk production losses in Latxa dairy sheep associated with small ruminant lentivirus infection. Prev. Veter Med. 2020;176:104886. doi: 10.1016/j.prevetmed.2020.104886. [DOI] [PubMed] [Google Scholar]
- 3.Blacklaws B.A. Small ruminant lentiviruses: Immunopathogenesis of visna-maedi and caprine arthritis and encephalitis virus. Comp. Immunol. Microbiol. Infect. Dis. 2012;35:259–269. doi: 10.1016/j.cimid.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Pritchard G.C., McConnel I. Maedi-Visna. In: Aitken I.D., editor. Diseases of Sheep. Blackwell Publishing; Oxford, UK: 2007. pp. 217–223. [Google Scholar]
- 5.Michiels R., Adjadj N.R., De Regge N. Phylogenetic Analysis of Belgian Small Ruminant Lentiviruses Supports Cross Species Virus Transmission and Identifies New Subtype B5 Strains. Pathogens. 2020;9:183. doi: 10.3390/pathogens9030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramírez H., Reina R., Amorena B., de Andrés D., Martínez H.A. Small ruminant Lentiviruses: Genetic variability, tropism and diagnosis. Viruses. 2013;5:1175–1207. doi: 10.3390/v5041175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gayo E., Polledo L., Balseiro A., Martínez C.P., García Iglesias M.J., Preziuso S., Rossi G., García Marín J.F. Inflammatory Lesion Patterns in Target Organs of Visna/Maedi in Sheep and their Significance in the Pathogenesis and Diagnosis of the Infection. J. Comp. Pathol. 2018;159:49–56. doi: 10.1016/j.jcpa.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Pinczowski P., Sanjosé L., Gimeno M., Crespo H., Glaria I., Amorena B., de Andrés D., Pérez M., Reina R., Luján L. Small Ruminant Lentiviruses in Sheep: Pathology and Tropism of 2 Strains Using the Bone Marrow Route. Vet. Pathol. 2017;54:413–424. doi: 10.1177/0300985816688742. [DOI] [PubMed] [Google Scholar]
- 9.De Andrés D., Klein D., Watt N., Berriatua E., Torsteinsdottir S., Blacklaws B., Harkiss G. Diagnostic tests for small ruminant lentiviruses. Veter Microbiol. 2005;107:49–62. doi: 10.1016/j.vetmic.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Larruskain A., Jugo B.M. Retroviral Infections in Sheep and Goats: Small Ruminant Lentiviruses and Host Interaction. Viruses. 2013;5:2043–2061. doi: 10.3390/v5082043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Echeverría I., De Miguel R., De Pablo-Maiso L., Glaria I., Benito A.A., De Blas I., De Andrés D., Luján L., Reina R. Multi-Platform Detection of Small Ruminant Lentivirus Antibodies and Provirus as Biomarkers of Production Losses. Front. Vet. Sci. 2020;7:182. doi: 10.3389/fvets.2020.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crespo H., Bertolotti L., Proffiti M., Cascio P., Cerruti F., Acutis P.L., de Andrés D., Reina R., Rosati S. Low proviral small ruminant lentivirus load as biomarker of natural restriction in goats. Vet. Microbiol. 2016;192:152–162. doi: 10.1016/j.vetmic.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Carrozza M.L., Mazzei M., Lacerenza D., Del Chiaro L., Giammarioli M., Marini C., Rutili D., Rosati S., Tolari F. Seroconversion against SU5 derived synthetic peptides in sheep experimentally infected with different SRLV genotypes. Vet. Microbiol. 2009;137:369–374. doi: 10.1016/j.vetmic.2009.01.032. [DOI] [PubMed] [Google Scholar]
- 14.Sigurdsson B., Grímsson H., Pálsson P. Maedi, a Chronic, Progressive Infection of Sheep’s Lungs. J. Infect. Dis. 1952;90:233–241. doi: 10.1093/infdis/90.3.233. [DOI] [PubMed] [Google Scholar]
- 15.Hutton B., Catalá-López F., Moher D. The PRISMA statement extension for systematic reviews incorporating network meta-analysis: PRISMA-NMA. Med. Clínica Engl. Ed. 2016;147:262–266. doi: 10.1016/j.medcle.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ. 2009;339:332–336. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson E.B. Probable Inference, the Law of Succession, and Statistical Inference. J. Am. Stat. Assoc. 1927;22:209. doi: 10.1080/01621459.1927.10502953. [DOI] [Google Scholar]
- 18.Wallace B.C., Dahabreh I.J., Trikalinos T.A., Lau J., Trow P., Schmid C.H. Closing the gap between methodologists and end-users: R as a computational back-end. J. Stat. Softw. 2012;49:1–15. doi: 10.18637/jss.v049.i05. [DOI] [Google Scholar]
- 19.Ayelet G., Roger F., Tibbo M., Tembely S. Survey of Maedi-Visna (MV) in Ethiopian Highland Sheep. Veter J. 2001;161:208–210. doi: 10.1053/tvjl.2000.0544. [DOI] [PubMed] [Google Scholar]
- 20.Bouljihad M., Leipold H.W. Ovine Lentiviral Infection (Maedi/Visna) in Morocco: A Serologic and Postmortem Survey. J. Veter Med. Ser. A. 1994;41:317–328. doi: 10.1111/j.1439-0442.1994.tb00098.x. [DOI] [PubMed] [Google Scholar]
- 21.Mahin L., Chadli M., Houwers D.J. A preliminary report on the occurrence of maedi-visna in sheep in Morocco. Veter Q. 1984;6:104. doi: 10.1080/01652176.1984.9693921. [DOI] [PubMed] [Google Scholar]
- 22.Belino E.D., Ezeifeka G.O. Maedi-visna antibodies in sheep and goats in Nigeria. Veter Rec. 1984;114:570. doi: 10.1136/vr.114.23.570-a. [DOI] [PubMed] [Google Scholar]
- 23.Albayrak H., Yazici Z., Okur-Gumusova S., Ozan E. Maedi-visna virus infection in Karayaka and Amasya Herik breed sheep from provinces in northern Turkey. Trop. Anim. Health Prod. 2011;44:939–941. doi: 10.1007/s11250-011-9996-9. [DOI] [PubMed] [Google Scholar]
- 24.Didugu H., Sagi S.B.R., Reddy C.E.N., Kishore K.N., Reddy M.V., Vishnu P.G. First report of Maedi-Visna and Caprine Arthritis- Encephalitis viruses in Krishna district, Andhra Pradesh, India. J. Anim. Res. 2016;6:177. doi: 10.5958/2277-940X.2016.00029.2. [DOI] [Google Scholar]
- 25.Muz D., Oǧuzoǧlu T.Ç., Rosati S., Reina R., Bertolotti L., Burgu I. First molecular characterization of visna/maedi viruses from naturally infected sheep in Turkey. Arch. Virol. 2013;158:559–570. doi: 10.1007/s00705-012-1518-1. [DOI] [PubMed] [Google Scholar]
- 26.Norouzi B., Taghavi Razavizadeh A., Azizzadeh M., Mayameei A., Najar V., Mashhadi N., Taghavi A., Dvm R. Serological study of small ruminant lentiviruses in sheep population of Khorasan-e-Razavi province in Iran. Vet. Res. Forum. 2015;6:245–249. [PMC free article] [PubMed] [Google Scholar]
- 27.Preziuso S., Or M.E., Giammarioli M., Kayar A., Feliziani F., Gönül R., Farneti S., Parkan Yaramiş Ç., Valente C., Cuteri V. Maedi-visna virus in Turkish sheep: A preliminary serological survey using ELISA tests. Turk. J. Vet. Anim. Sci. 2010;34:289–293. [Google Scholar]
- 28.Tolari F., Al-Ramadneh W., Mazzei M., Carrozza M.L., Forzan M., Bandecchi P., Grego E., Rosati S. Small ruminant lentiviruses in Jordan: Evaluation of sheep and goat serological response using recombinant and peptide antigens. Trop. Anim. Health Prod. 2013;45:1335–1340. doi: 10.1007/s11250-013-0366-7. [DOI] [PubMed] [Google Scholar]
- 29.Zhang K s.h.a.n., He J j.u.n., Liu Y j.i.e., Shang Y j.u.n., Liu X.T. A seroprevalence survey of maedi-visna among twenty-four ovine floks from twelve regions of China. J. Integr. Agric. 2013;12:2321–2323. doi: 10.1016/S2095-3119(13)60380-9. [DOI] [Google Scholar]
- 30.Azkur A.K., Gazyagci S., Aslan M.E. Serological and Epidemiological Investigation of Bluetongue, Maedi-Visna and Caprine Arthritis-Encephalitis Viruses in Small Ruminant in Kirikkale District in Turkey. Kafkas Universitesi Vet. Fakulltesi Derg. 2011;17:803–808. [Google Scholar]
- 31.Giangaspero M., Osawa T., Orusa R., Frossard J.-P., Naidu B., Robetto S., Tatami S., Takagi E., Moriya H., Okura N., et al. Epidemiological survey for visna-maedi among sheep in northern prefectures of Japan. Veter Ital. 2011;47:437–451. [PubMed] [Google Scholar]
- 32.Mosa A.H., Zenad M.M. First Molecular Detection of Maedi-Visna Virus in Awassi Sheep of Middle Iraq Regions. Bulg. J. Vet. Med. 2020 doi: 10.15547/bjvm.2020-0069. [DOI] [Google Scholar]
- 33.Tabet E., Tlaige R., El Hage J., Abi-Rizk A. The occurrence of Maedi-Visna Virus in Lebanon. Volume 36. O.I.E (World Organisation for Animal Health); Paris, France: 2017. [DOI] [PubMed] [Google Scholar]
- 34.Mahmood F., Khan A., Khan M.Z., Hussain R., Gul S.T., Siddique A.B. Pathological and molecular based study of naturally occurring lentivirus infection. Pak. Vet. J. 2012;32:511–514. [Google Scholar]
- 35.Giangaspero M., Vanopdenbosch E., Nishikawa H., Tabbaa D. Prevalence of antibodies against respiratory viruses (parainfluenza virus type 3, respiratory syncytial virus, reovirus and adenovirus) in relation to productivity in Syrian Awassi sheep. Trop. Anim. Health Prod. 1997;29:83–91. doi: 10.1007/BF02632323. [DOI] [PubMed] [Google Scholar]
- 36.Alba A., Allepuz A., Serrano E., Casal J. Seroprevalence and spatial distribution of maedi-visna virus and pestiviruses in Catalonia (Spain) Small Rumin. Res. 2008;78:80–86. doi: 10.1016/j.smallrumres.2008.05.004. [DOI] [Google Scholar]
- 37.Lago N., Lopez C., Panadero R., Cienfuegos S., Pato J., Prieto A., Díaz P., Mourazos N., Fernández G. Seroprevalence and risk factors associated with Visna/Maedi virus in semi-intensive lamb-producing flocks in northwestern Spain. Prev. Veter Med. 2012;103:163–169. doi: 10.1016/j.prevetmed.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 38.León L., Prats J. Encuesta serológica de la infección por el virus Maedi/Visna en la población ovina de Moratalla (Murcia); Proceedings of the XXI Jornadas Científicas de la Sociedad Española de Ovinotecnia y Caprinotecnia (SEOC); Murcia, Spain. 3–5 October 1996; pp. 119–127. [Google Scholar]
- 39.Olech M., Osiński Z., Kuźmak J. Bayesian estimation of seroprevalence of small ruminant lentiviruses in sheep from Poland. Prev. Veter Med. 2017;147:66–78. doi: 10.1016/j.prevetmed.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Rémond M., Larenaudie B. Maladie Visna-Maedi en France: Bilan d’une enquête sérologique et perspectives d’eradication. Rev. Sci. Technol. Off. Int. Epiz. 1982;1:429–433. doi: 10.20506/rst.1.2.72. [DOI] [PubMed] [Google Scholar]
- 41.Pérez M., Biescas E., de Andrés X., Leginagoikoa I., Salazar E., Berriatua E., Reina R., Bolea R., de Andrés D., Juste R.A., et al. Visna/maedi virus serology in sheep: Survey, risk factors and implementation of a successful control programme in Aragón (Spain) Vet. J. 2010;186:221–225. doi: 10.1016/j.tvjl.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 42.Ritchie C., Davies I., Smith R. Maedi Visna Seroprevalence Survey 2010. [(accessed on 18 December 2020)];2012 Available online: https://meatpromotion.wales/images/resources/Funding_bodies_report_MV_seroprevalence_survey_2010_final_version_14.6.12.pdf.
- 43.Savic S., Žekić Stošić M., Bugarski D., Marčić D., Milovanović A., Potkonjak A. Prevalence of Small Ruminant Lentivirus Infections In Sheep and Goats in Some Regions of Vojvodina Province. Arch. Vet. Med. 2020;13:29–40. doi: 10.46784/e-avm.v13i1.53. [DOI] [Google Scholar]
- 44.Schaller P., Vogt H.R., Strasser M., Nettleton P.F., Peterhans E., Zanoni R. Seroprevalence of Maedi-Visna and Border Disease in Switzerland. Bord. Dis. Switz. 2000;142:145–153. [PubMed] [Google Scholar]
- 45.Sihvonen L., Nuotio L., Rikula U., Hirvelä-Koski V., Kokkonen U.-M. Preventing the spread of maedi-visna in sheep through a voluntary control programme in Finland. Prev. Veter Med. 2000;47:213–220. doi: 10.1016/S0167-5877(00)00162-8. [DOI] [PubMed] [Google Scholar]
- 46.Straub O.C. Maedi–Visna virus infection in sheep. History and present knowledge. Comp. Immunol. Microbiol. Infect. Dis. 2004;27:1–5. doi: 10.1016/S0147-9571(02)00078-4. [DOI] [PubMed] [Google Scholar]
- 47.Berriatua E., Álvarez V., Extramiana B., González L., Daltabuit M., Juste R. Transmission and control implications of seroconversion to Maedi-Visna virus in Basque dairy-sheep flocks. Prev. Vet. Med. 2003;60:265–279. doi: 10.1016/S0167-5877(03)00163-6. [DOI] [PubMed] [Google Scholar]
- 48.Cana A., Taylor N., Honhold N., Gjinovci V., Osmani A., Nardelli S., Murati B., Spahiu J., Mehmetukaj D., Alishani M. Sero-prevalence survey of small ruminant lentivirus (SRLV) infections in Kosovo. Kafkas Univ. Vet. Fak. Derg. 2020;26:1–8. [Google Scholar]
- 49.Caporale V.P., Foglini A., Lelli R., Mantovani A., Nannini D., Simoni P. Preliminary observations on the presence of visna-maedi in Italy. Vet. Res. Commun. 1983;6:31–35. doi: 10.1007/BF02214893. [DOI] [PubMed] [Google Scholar]
- 50.Espi-Felgueroso A., Prieto-Martín J.M., Álvarez-Martínez J.M., Pereira-Bueno J. Situación sanitaria del ovino en Asturias: Seroprevalencia de la Enfermedad de la Frontera, Maedi-Visna, Aborto Enzoótico, Agalaxia Contagiosa, Fiebre Q y Leptospirosis; Proceedings of the XXVI Jornadas Científicas y V Internacionales de la Sociedad Española de Ovinotecnia y Caprinotecnia; Seville, Spain. 20–22 September 2001. [Google Scholar]
- 51.Huttner K., Seelmann M., Feldhusen F. Prevalence and risk factors for Maedi-Visna in sheep farms in Mecklenburg-Western-Pomerania. Berl. Munch. Tierarztl. Wochenschr. 2010;123:463–467. [PubMed] [Google Scholar]
- 52.Junkuszew A., Dudko P., Bojar W., Olech M., Osiński Z., Gruszecki T.M., Kania M.G., Kuźmak J., Czerski G. Risk factors associated with small ruminant lentivirus infection in eastern Poland sheep flocks. Prev. Veter Med. 2016;127:44–49. doi: 10.1016/j.prevetmed.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 53.Karanikolaou K., Angelopoulou K., Papanastasopoulou M., Koumpati-Artopiou M., Papadopoulos O., Koptopoulos G. Detection of small ruminant lentiviruses by PCR and serology tests in field samples of animals from Greece. Small Rumin. Res. 2005;58:181–187. doi: 10.1016/j.smallrumres.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 54.Michiels R., Van Mael E., Quinet C., Welby S., Cay A.B., De Regge N. Seroprevalence and risk factors related to small ruminant lentivirus infections in Belgian sheep and goats. Prev. Veter Med. 2018;151:13–20. doi: 10.1016/j.prevetmed.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 55.Barták P., Šimek B., Václavek P., Čurn V., Plodková H., Tonka T., Farková B., Vernerová K., Vejčík A. Genetic characterisation of small ruminant lentiviruses in sheep and goats from the Czech Republic. Acta Vet. Brno. 2018;87:19–26. doi: 10.2754/avb201887010019. [DOI] [Google Scholar]
- 56.Hönger D., Leitold B., Schuller W. Serological studies of antibodies against maedi-visna virus in sheep in Austria. Berl Munch Tierarztl Wochenschr. 1990;103:39–41. [PubMed] [Google Scholar]
- 57.Arsenault J., Dubreuil P., Girard C., Simard C., Bélanger D. Maedi-visna impact on productivity in Quebec sheep flocks (Canada) Prev. Veter Med. 2003;59:125–137. doi: 10.1016/S0167-5877(03)00086-2. [DOI] [PubMed] [Google Scholar]
- 58.Campbell J.R., Menzies P.I., Waltner-Toews D., Walton J.S., Buckrell B.C., Thorsen J. The seroprevalence of maedi-visna in Ontario sheep flocks and its relationship to flock demographics and management practices. Can. Veter J. 1994;35:39–44. [PMC free article] [PubMed] [Google Scholar]
- 59.Cutlip R.C., Lehmkuhl H.D., Sacks J.M., Weaver A.L. Seroprevalence of ovine progressive pneumonia virus in sheep in the United States as assessed by analyses of voluntarily submitted samples. Am. J. Veter Res. 1992;53:976–979. [PubMed] [Google Scholar]
- 60.Fournier D., Campbell J.R., Middleton D.M. Prevalence of maedi-visna infection in culled ewes in Alberta. Can. Veter J. 2006;47:460–466. [PMC free article] [PubMed] [Google Scholar]
- 61.Gerstner S., Adamovicz J.J., Duncan J.V., Laegreid W.W., Marshall K.L., Logan J.R., Schumaker B.A. Prevalence of and risk factors associated with ovine progressive pneumonia in Wyoming sheep flocks. J. Am. Veter Med Assoc. 2015;247:932–937. doi: 10.2460/javma.247.8.932. [DOI] [PubMed] [Google Scholar]
- 62.Shuaib M., Green C., Rashid M., Duizer G., Whiting T.L. Herd risk factors associated with sero-prevalence of Maedi-Visna in the Manitoba sheep population. Can. Veter J. 2010;51:385–390. [PMC free article] [PubMed] [Google Scholar]
- 63.Simard C., Morley R.S. Seroprevalence of maedi-visna in Canadian sheep. Can. J. Veter Res. 1991;55:269–273. [PMC free article] [PubMed] [Google Scholar]
- 64.Uzcanga M.G. Estudio Epidemiológico de Maedi-Visna en Ovinos de la Región de los Tuxtlas del Estado de Veracruz. [(accessed on 10 December 2020)];2015 Available online: https://cdigital.uv.mx/handle/1944/49707.
- 65.Centers for Epidemiology and Animal Health Ovine Progressive Pneumonia: Awareness, Management, and Seroprevalance; APHIS Info Sheet. [(accessed on 28 November 2020)]; Available online: https://www.aphis.usda.gov/animal_health/nahms/sheep/downloads/sheep01/Sheep01_is_OPP.pdf.
- 66.Heinrichs R., Wilkins W., Schroeder G., Campbell J. Prevalence of Maedi-visna in Saskatchewan sheep. Can. Veter J. 2017;58:183–186. [PMC free article] [PubMed] [Google Scholar]
- 67.Villagra-Blanco R., Dolz G., Solórzano-Morales A., Alfaro A., Montero-Caballero D., Romero-Zúñiga J.J. Presence of Maedi-Visna in Costa Rican sheep flocks. Small Rumin. Res. 2015;124:132–136. doi: 10.1016/j.smallrumres.2015.01.010. [DOI] [Google Scholar]
- 68.Alves J.R.A., Limeira C.H., Lima G.M.S., Pinheiro R.R., Alves F.S.F., Dos Santos V.W.S., De Azevedo S.S., Alves C.J. Epidemiological characterization and risk factors associated with lentiviral infection of small ruminants at animal fairs in the semiarid Sertão region of Pernambuco, Brazilian semiarid. Semin. Ciênc. Agrárias. 2017;38:1875–1886. doi: 10.5433/1679-0359.2017v38n4p1875. [DOI] [Google Scholar]
- 69.Da Costa L.S.P., de Lima P.P., Callado A.K.C., do Nascimento S.A., de Castro R.S. Lentivírus de pequenos ruminantes em ovinos Santa Inês: Isolamento, identificação pela PCR e inquérito sorológico no estado de Pernambuco. Arq. Inst. Biol. 2007;74:11–16. [Google Scholar]
- 70.Martinez P.M., Costa J.N., De Souza T.S., Costa Neto A.O., Pinheiro R.R. Revalência sorológica da Maedi-visna em rebanhos ovinos da microrregião de Juazeiro - Bahia por meio do teste de Imunodifusão em gel de ágar. Ciênc. Anim. Bras. 2011;12:322–329. doi: 10.5216/cab.v12i2.4454. [DOI] [Google Scholar]
- 71.Mendonça C.E.D., Barros S.L.B., Mendonça M.A.D., Guimarães V.A.A., Pinheiro R.R. Ocorrência de anticorpos contra o vírus Maedi-Visna em ovinos Santa Inês, no Estado de Sergipe, Brasil. Arq. Inst. Biol. 2013;80:346–351. doi: 10.1590/S1808-16572013000300013. [DOI] [Google Scholar]
- 72.Robles C.A., Layana J.E., Cabrera R., Raffo F., Cultlip R. Estudio serológico retrospectivo de Maedi (Neumonía Progresiva) en ovinos y de Artritis- Encefalitis en caprinos de Patagonia, Argentina. Rev. Med. Vet. 2003;84:96–99. [Google Scholar]
- 73.Trezeguet M.Á., Suárez M.F., Barral L.E., Periolo F., Maidana C.E., Farías P.C., Rodríguez C.E., Teresa R. Situación epidemiológica de maedi-visna y artritis encefalitis caprina en la Argentina. Sitio Argent. Prod. Anim. 2013;1:1–11. [Google Scholar]
- 74.Vinha K.T., da Silva T.I.B. Seropositivity for Maedi-Visna virus in sheep in Porto Acre city—Western Amazon, Brazil. Ciênc. Anim. Bras. 2020;21:e59173. doi: 10.1590/1809-6891v21e-59173. [DOI] [Google Scholar]
- 75.Lombardi A.L., Nogueira A.H.C., Feres F.C., Paulo H.P., Castro R.S., Feitosa F.L.F., Cadioli F.A., Peiro J.R., Perri S.H.V., Lima V.F.M., et al. Occurrence of Maedi-visna in sheep from Aracatuba region—SP—Brazil. Arq. Bras. Med. Vet. Zootec. 2009;61:1434–1437. doi: 10.1590/S0102-09352009000600025. [DOI] [Google Scholar]
- 76.Carrozza M.-L., Niewiadomska A.-M., Mazzei M., Abi-Said M.R., Hué S., Singer J.B., Hughes J., Gifford R.J. An investigation into the origins and history of pandemic small ruminant lentivirus infection. bioRxiv. 2018 doi: 10.1101/236117. [DOI] [Google Scholar]
- 77.Houwers D.J. Maedi-Visna and Related Diseases. Springer; Boston, MA, USA: 1990. Economic Importance, Epidemiology and Control; pp. 83–117. [Google Scholar]
- 78.Pálsson P.A. Maedi-Visna and Related Diseases. Springer; Boston, MA, USA: 1990. Maedi-Visna. History and Clinical Description; pp. 3–17. [Google Scholar]
- 79.Álvarez V., Arranz J., Daltabuit-Test M., Leginagoikoa I., Juste R.A., Amorena B., de Andrés D., Luján L.L., Badiola J.J., Berriatua E. Relative contribution of colostrum from Maedi-Visna virus (MVV) infected ewes to MVV-seroprevalence in lambs. Res. Vet. Sci. 2005;78:237–243. doi: 10.1016/j.rvsc.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 80.Kilgour R.J., Waterhouse T., Dwyer C.M., Ivanov I.D. The Welfare of Sheep. Springer; Dordrecht, The Netherlands: 2008. Farming Systems for Sheep Production and Their Effect on Welfare; pp. 213–265. [Google Scholar]
- 81.Pulina G., Milán M., Lavín M., Theodoridis A., Morin E., Capote J., Thomas D., Francesconi A., Caja G. Invited review: Current production trends, farm structures, and economics of the dairy sheep and goat sectors. J. Dairy Sci. 2018;101:6715–6729. doi: 10.3168/jds.2017-14015. [DOI] [PubMed] [Google Scholar]
- 82.Morris S.T. Advances in Sheep Welfare. Elsevier; Amsterdam, The Netherlands: 2017. Overview of sheep production systems; pp. 19–35. [Google Scholar]
- 83.Gilbert M., Nicolas G., Cinardi G., Van Boeckel T.P., Vanwambeke S.O., Wint G.R.W., Robinson T.P. Global distribution data for cattle, buffaloes, horses, sheep, goats, pigs, chickens and ducks in 2010. Sci. Data. 2018;5:1–11. doi: 10.1038/sdata.2018.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sheep Livestock Systems. [(accessed on 12 January 2021)];Food and Agriculture Organization of the United Nations. Available online: http://www.fao.org/livestock-systems/global-distributions/sheep/en/
- 85.Global Sheep Distribution in 2010 (5 minutes of arc)—Gridded Livestock of the World—Latest—2010 (GLW 3) [(accessed on 17 January 2021)]; doi: 10.7910/DVN/BLWPZN. Available online: https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/BLWPZN. [DOI]
- 86.Singh C., Köhler-Rollefson I., Box P., Pali D. Sheep pastoralism in Rajasthan: Still a viable livelihood option?; Proceedings of the State-Level Workshop of The LIFE Initiative Local Livestock for Empowerment of Rural People; India. 1 May 2005; [(accessed on 15 January 2021)]. Available online: http://www.pastoralpeoples.org/wp-content/uploads/2020/01/sheep_workshop.pdf. [Google Scholar]
- 87.Holden S., Shiferaw B. Policies for sustainable land management in the highlands of Ethiopia; Development paths and policies for sustainable land management in Andit Tid, North Shewa: An exploration. In proceedings of the Socio-Economics and Policy International Livestock Research Institute; Addis Ababa, Ethiopia. 22–23 May 2000; [(accessed on 15 January 2021)]. Available online: https://land.igad.int/index.php/documents-1/countries/ethiopia/rural-development-1/343-policies-for-sustainable-land-management-in-the-highlands-of-ethiopia/file. [Google Scholar]
- 88.Mekoya A., Dar B., Yami A., Habtemariam M. Management of Traditional Sheep Production in Lallomamma Mider Wereda, North Shoa, Amhara. [(accessed on 17 January 2021)]; Available online: https://agris.fao.org/agris-search/search.do?recordID=ET2002000017.
- 89.Ritchie C., Hosie B. Concern over maedi visna breakdowns. Veter Rec. 2014;175:50–51. doi: 10.1136/vr.g4522. [DOI] [PubMed] [Google Scholar]
- 90.Synge B.A., Ritchie C.M. Elimination of small ruminant lentivirus infection from sheep flocks and goat herds aided by health schemes in Great Britain. Veter Rec. 2010;167:739–743. doi: 10.1136/vr.c5853. [DOI] [PubMed] [Google Scholar]
- 91.Barquero N., Gomez-Lucia E., Arjona A., Toural C., Heras A., Fernández-Garayzábal J.F., Quiteria J.A., Doménech A. Investigation of risk factors associated with infections caused by small ruminant lentiviruses. Bull. Veter Inst. Pulawy. 2013;57:473–478. doi: 10.2478/bvip-2013-0082. [DOI] [Google Scholar]
- 92.Leginagoikoa I., Daltabuit-Test M., Álvarez V., Arranz J., Juste R.A., Amorena B., de Andrés D., Luján L.L., Badiola J.J., Berriatua E. Horizontal Maedi-Visna virus (MVV) infection in adult dairy-sheep raised under varying MVV-infection pressures investigated by ELISA and PCR. Res. Vet. Sci. 2006;80:235–241. doi: 10.1016/j.rvsc.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 93.Glaria I., Reina R., Ramírez H., de Andrés X., Crespo H., Jauregui P., Salazar E., Luján L., Pérez M.M., Benavides J., et al. Visna/Maedi virus genetic characterization and serological diagnosis of infection in sheep from a neurological outbreak. Vet. Microbiol. 2012;155:137–146. doi: 10.1016/j.vetmic.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 94.Saman E., Van Eynde G., Lujan L., Extramiana B., Harkiss G., Tolari F., Gonzalez L., Amorena B., Watt N., Badiola J. A New Sensitive Serological Assay for Detection of Lentivirus Infections in Small Ruminants. Clin. Diagn. Lab. Immunol. 1999;6:734–740. doi: 10.1128/CDLI.6.5.734-740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leginagoika I., Juste R.A., Barandika J., Amorena B., De Andrés D., Luján L., Badiola J., Berriatua E. Extensive rearing hinders Maedi-Visna Virus (MVV) infection in sheep. Vet. Res. 2006;37:767–778. doi: 10.1051/vetres:2006034. [DOI] [PubMed] [Google Scholar]
- 96.Álvarez V., Daltabuit-Test M., Arranz J., Leginagoikoa I., Juste R.A., Amorena B., de Andrés D., Luján L.L., Badiola J.J., Berriatua E. PCR detection of colostrum-associated Maedi-Visna virus (MVV) infection and relationship with ELISA-antibody status in lambs. Res. Vet. Sci. 2006;80:226–234. doi: 10.1016/j.rvsc.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 97.De Andrés X., Ramírez H., Bertolotti L., San Román B., Glaria I., Crespo H., Jáuregui P., Minguijón E., Juste R., Leginagoikoa I., et al. An insight into a combination of ELISA strategies to diagnose small ruminant lentivirus infections. Vet. Immunol. Immunopathol. 2013;152:277–288. doi: 10.1016/j.vetimm.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 98.Brinkhof J., Houwers D., Moll L., Dercksen D., Van Maanen C. Diagnostic performance of ELISA and PCR in identifying SRLV-infected sheep and goats using serum, plasma and milk samples and in early detection of infection in dairy flocks through bulk milk testing. Veter Microbiol. 2010;142:193–198. doi: 10.1016/j.vetmic.2009.09.060. [DOI] [PubMed] [Google Scholar]
- 99.Chassalevris T., Chaintoutis S.C., Apostolidi E.D., Giadinis N.D., Vlemmas I., Brellou G.D., Dovas C.I. A highly sensitive semi-nested real-time PCR utilizing oligospermine-conjugated degenerate primers for the detection of diverse strains of small ruminant lentiviruses. Mol. Cell. Probes. 2020;51:101528. doi: 10.1016/j.mcp.2020.101528. [DOI] [PubMed] [Google Scholar]
- 100.Ramírez H., Echeverría I., Benito A.A., Glaria I., Benavides J., Pérez V., de Andrés D., Reina R. Accurate Diagnosis of Small Ruminant Lentivirus Infection Is Needed for Selection of Resistant Sheep through TMEM154 E35K Genotyping. Pathogens. 2021;10:83. doi: 10.3390/pathogens10010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cardinaux L., Zahno M.L., Deubelbeiss M., Zanoni R., Vogt H.R., Bertoni G. Virological and phylogenetic characterization of attenuated small ruminant lentivirus isolates eluding efficient serological detection. Vet. Microbiol. 2013;162:572–581. doi: 10.1016/j.vetmic.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 102.Molaee V., Eltanany M., Lühken G. First survey on association of TMEM154 and CCR5 variants with serological maedi-visna status of sheep in German flocks. Vet. Res. 2018;49:1–10. doi: 10.1186/s13567-018-0533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Field N., Cohen T., Struelens M.J., Palm D., Cookson B., Glynn J.R., Gallo V., Ramsay M., Sonnenberg P., MacCannell D., et al. Strengthening the Reporting of Molecular Epidemiology for Infectious Diseases (STROME-ID): An extension of the STROBE statement. Lancet Infect. Dis. 2014;14:341–352. doi: 10.1016/S1473-3099(13)70324-4. [DOI] [PubMed] [Google Scholar]
- 104.PRISMA Transparent Reporting of Systematic Reviews and Meta-Analyses—PRISMA Statement Flow Diagram. [(accessed on 13 December 2020)]; Available online: http://www.prisma-statement.org/PRISMAStatement/FlowDiagram.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available in the scientific publications listed in the Reference section.