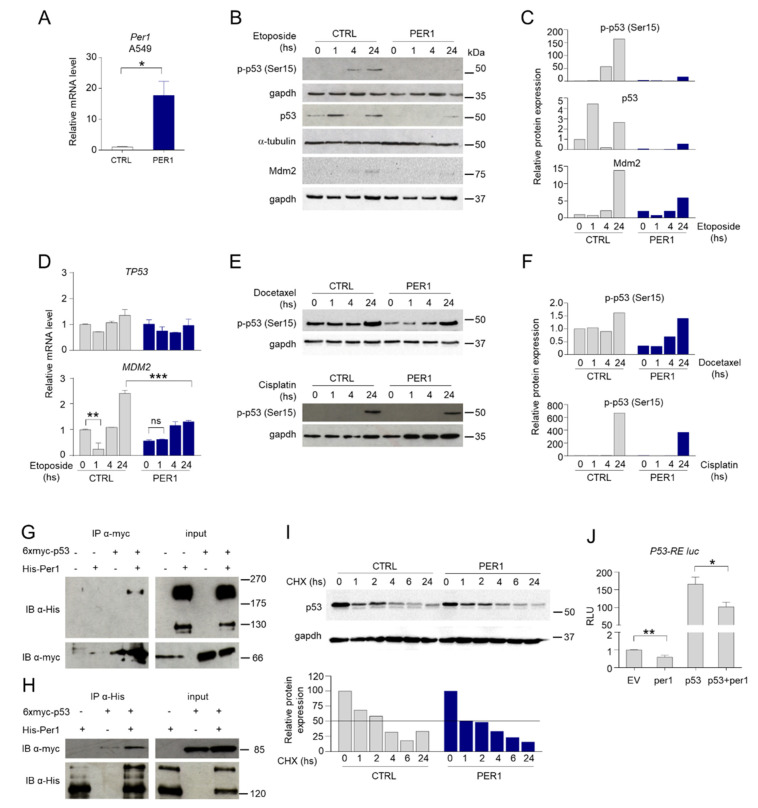
Figure 1.
PER1 modulation of p53 expression and activity. (A) Control of PER1-overexpressing clones in A549 cells, evaluated by qPCR. A549 cells were transfected with PER1 or a control plasmid (CTRL, pEGFP N1) and positive clones were selected with G418 treatment. Bars represent average ± SD (n = 2). (*) p < 0.05. (B) Levels of p-p53 (Ser15), total p53 and Mdm2 following treatment with etoposide for the indicated time in A549 cells stably overexpressing PER1 or CTRL vector, evaluated by Western blotting (WB). Gapdh and α-tubulin were used as loading controls. (C) Densitometry for p-p53 (Ser15), total p53 and Mdm2 expression levels relative to (B). (D) TP53 and MDM2 gene expression levels in A549 cells stably overexpressing PER1 or CTRL vector treated as in 2B. Bars represent average ± SD (n = 2). (**) p < 0.01, (***) p < 0.001. (E) Levels of p-p53 (Ser15) following treatment with docetaxel or cisplatin for the indicated time in A549 cells stably overexpressing PER1 or CTRL vector, evaluated by WB. Gapdh was used as loading controls. (F) Densitometry for p-p53 (Ser15) expression levels relative to (E). (G–H) Immunoprecipitation (IP) between PER1 and p53. HEK-293 cells were cotransfected with myc-p53 and His-PER1. In (G) p53 was immunoprecipitated with anti-myc antibody. In (H) PER1 was immunoprecipitated with anti-His antibody. Co-immunoprecipitated proteins and total cell lysate (input) were analyzed by WB and revealed with anti-His and anti-myc antibodies. (I) Expression of p53 in A549 cells overexpressing PER1 or a control plasmid (CTRL) and treated with cycloheximide (CHX, 100 μM) for the indicated times. Gapdh was used as loading control. Densitometry for p53 expression levels is shown on the bottom. (J) Evaluation of the activation of p53-RE-Luc by luciferase assay in HEK-293 cells transfected with the indicated plasmids (ev = empty vector). Levels of expression are relative to the control transfected with empty vector, after normalization for transfection efficiency through beta-galattosidase assay. Bars represent average of RLU (relative luciferase units) ± SD (n = 3). (*) p < 0.05, (**) p < 0.01.