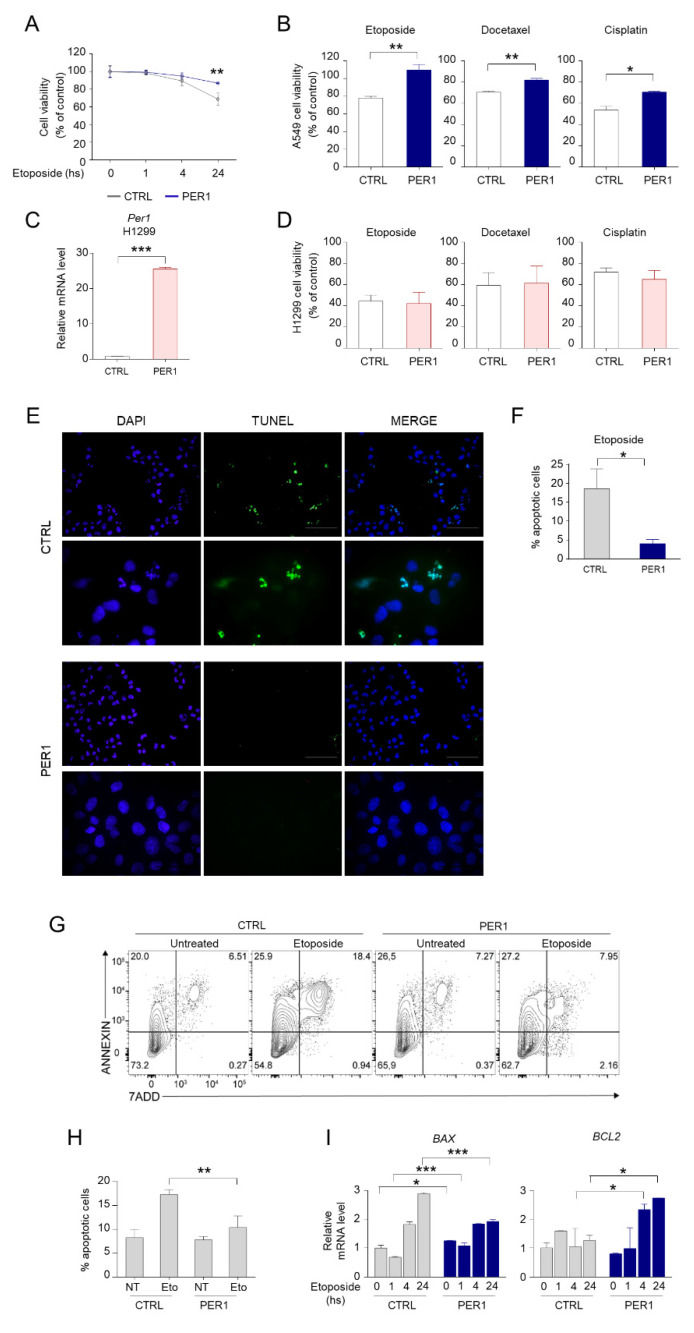
Figure 2.
PER1 downregulation of apoptosis. (A) Cell viability following treatment with etoposide for the indicated time in A549 cells overexpressing PER1 or CTRL vector, measured by MTT assay and expressed as % of control untreated cells. Bars represent average ± SD (n = 3). (**) p < 0.01. (B) Cell viability following treatment with etoposide, docetaxel or cisplatin for 24 h in A549 cells overexpressing PER1 or CTRL vector, expressed as % of control untreated cells. Bars represent average ± SD (n = 3). (*) p < 0.05, (**) p < 0.01. (C) Control of PER1-overexpressing clones in H1299 cells, evaluated by qPCR. H1299 cells were transfected with PER1 or a control plasmid (CTRL, pEGFP N1) and positive clones were selected with G418 treatment. Bars represent average ± SD (n = 2). (***) p < 0.001. (D) Cell viability following treatment with etoposide, docetaxel or cisplatin for 24 h in H1299 cells overexpressing PER1 or CTRL vector, expressed as % of control untreated cells. Bars represent average ± SD (n = 3). (E) Representative image of TUNEL staining of A549 cells overexpressing PER1 or CTRL vector treated with etoposide. Nuclei were counterstained with DAPI. 40× and 100× magnification (scale bars, 100 and 50 μm, respectively). (F) Percentage of apoptotic cells in CTRL and PER1 cells obtained by TUNEL-positive cell count. Bars represents means ± SD (n = 5 fields of observation from three samples each group). (G) Measure of apoptotic cells in A549 cells overexpressing PER1 or CTRL untreated or treated with etoposide, analyzed by flow cytometry. (H) Percentage of apoptotic cells in CTRL and PER1 cells, obtained by flow cytometry analysis. Bars represents average ± SD (n = 3). (I) BAX and BCL2 gene expression levels in A549 cells overexpressing PER1 or CTRL vector and treated with etoposide for the indicated time. Bars represent average ± SD (n = 2). (*) p < 0.05, (**) p < 0.01, (***) p < 0.001.