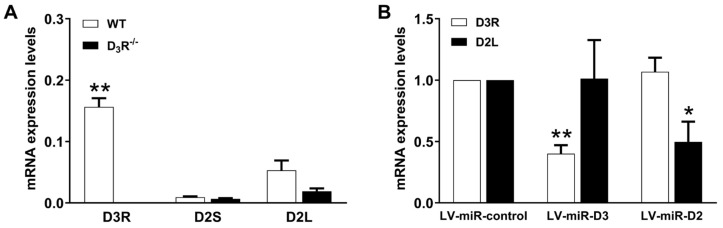
Figure 4.
(A) Levels of mRNA encoding D3R and D2R (short- and long-splice variants, D2S, D2L) in aortic segments from wild-type (WT) and D3R−/− mice. Total RNA was reverse transcribed; the corresponding cDNA was quantitated by real-time PCR. Levels of mRNA are normalized against the housekeeping gene GAPDH. Each column (with vertical bars representing standard errors) represents the average from 7–15 different RNA samples. Two-way ANOVA, (genotype: F(1,59) = 75.2, p < 0.0001; mRNA expr.: F(2,59) = 37.29, p < 0.0001; genotype × mRNA expr. F(2,59) = 46.88, p = 0.0001) and Bonferroni’s test (** p < 0.01 vs. D2S and D2L). (B) Levels of mRNA encoding D3R and D2R in aortic segments from wild-type treated with lentiviral vectors, delivering synthetic miRNA (LV-miR-control, scrambled sequence; LV-miR-D3, targeting D3R; LV-miR-D2, targeting D2R). Total RNA was reverse transcribed; the corresponding cDNA was quantitated by real-time PCR. Levels of mRNA are normalized against the housekeeping gene GAPDH and the mRNA receptor expression in LV-miR-control. Each column (with vertical bars representing standard errors) represents the average from 5–9 different RNA samples. Two-way ANOVA, (LV-miR: F(2,30) = 2.21 p = 0.12; mRNA expr.: F(1,30) = 0.01, p = 0.90; LV-miR × mRNA expr. F(2,30) = 9.39, p = 0.0007) and Bonferroni’s test (* p < 0.05, ** p < 0.01 vs. LV-miR-control).