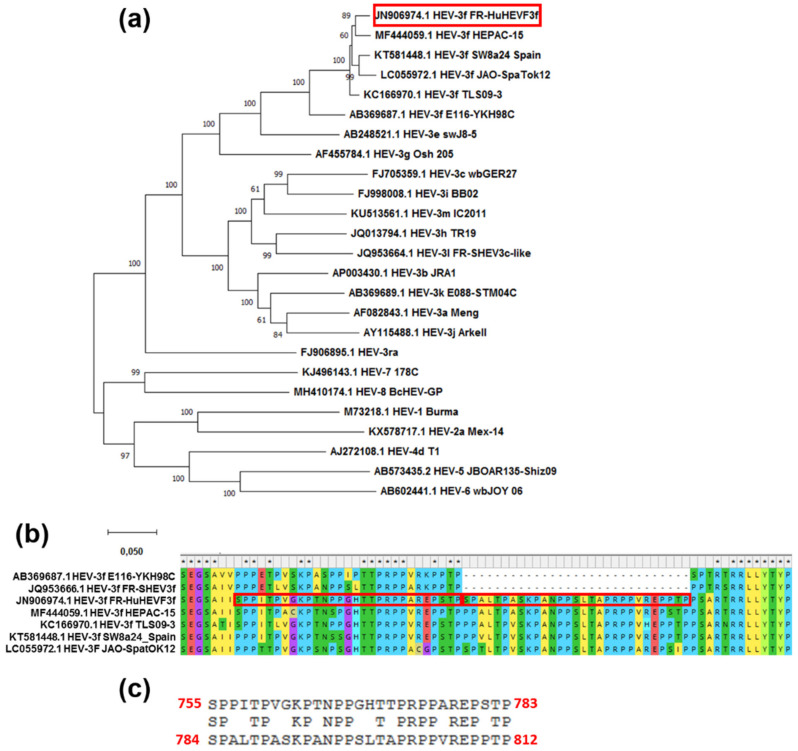
Figure 5.
Duplication of a 29 amino acid sequence within the HVR of FR-HuHEVF3f ORF1. (a) Phylogenetic tree of 25 HEV complete or nearly complete sequences. This analysis includes FR-HuHEVF3f, different referenced genotypes of HEV (HEV-1 to -8) and subtypes of HEV-3 as well the 4 HEV-3f strains shown to be the most closely related to FR-HuHEVF3f in a blast analysis (HEPAC-15, TLS09-3, SW8a24 Spain and JAO-SpaTok12). The tree was constructed by using the Maximum Likelihood method and Tamura-Nei model based on the Clustal W alignment in MEGA X software. Bootstrap values were obtained from 1000 replicates. The GenBank database accession number of these complete sequences is indicated. FR-HuHEVF3f is indicated by a red box. Scale bar represents nucleotide substitutions per site. (b) Alignment of FR-HuHEVF3f with other HEV-3f strains revealed the presence of a duplicated 29 amino acid region in the hypervariable region (HVR) domain of ORF1 as indicated by red boxes. (c) Alignment of the duplicated sequence. The amino acid position within FR-HuHEVF3f ORF1 is indicated in red.