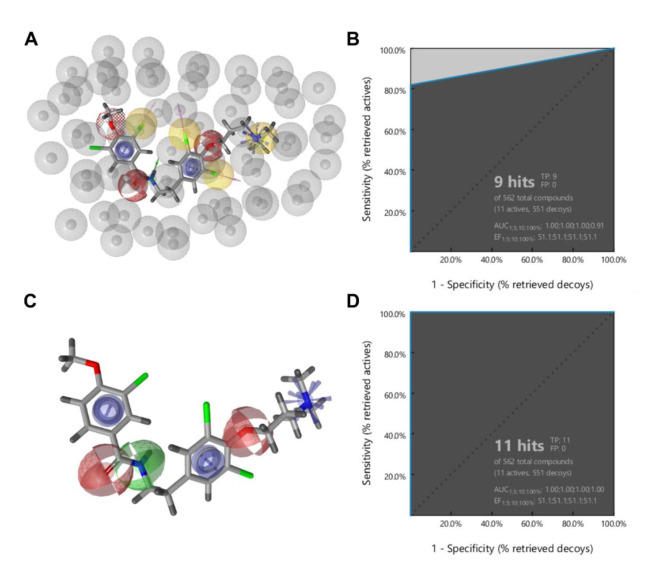
Figure 7.
(A) Initial ligand-based pharmacophore model for KV10.1 inhibitors based on a library of purpurealidin analogues. Common chemical features of the aligned molecules include 4 hydrophobic features (yellow spheres), 3 hydrogen bond acceptors (red spheres), 1 hydrogen bond donor (a green arrow), 2 aromatic features (blue discs), 1 positive ionisable feature (blue star), 3 halogen bond features (pink arrows), and exclusion volumes (grey spheres) that define restricted regions based on the shape of the aligned molecules. Optional features are marked as dashed. (C) Final ligand-based pharmacophore model. Common chemical features of the aligned molecules include 2 hydrogen bond acceptors (red spheres), 1 hydrogen bond donor (a green sphere), 2 aromatic features (blue discs), and 1 positive ionisable feature (blue star). Grey spheres for exclusion volumes are not shown. Resulting ROC plot (curve shown in blue) for (B) initial and (D) final LBPM from virtually screening 562 compounds (11 KV10.1 active compounds and 551 generated decoys) with the ligand-based pharmacophore model. TP = true positives; FP = false positives; AUC = area under the curve; EF = enrichment factor.