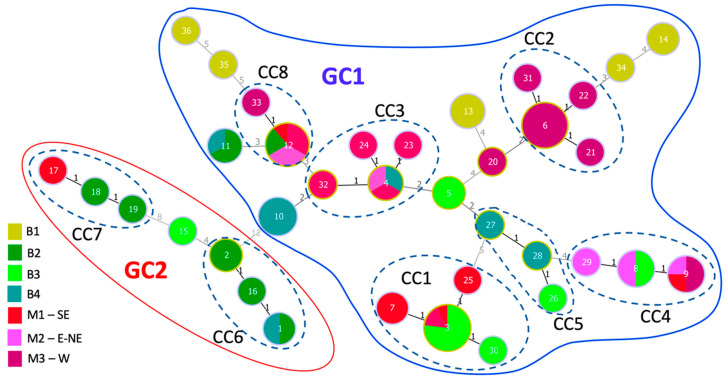
Figure 2.
Categorical minimum spanning tree from the VNTR-rep dataset using the goeBURST and Euclidian algorithms. Numbers in circles identify the 36 MLVA haplotypes. The circle sizes are proportional to the number of strains per haplotype, with the smallest circles (e.g., no. 17 in CC7) corresponding to one strain and the largest circle (no. 3 in CC1) corresponding to 13 strains. Colours indicate the country and region of origin, with four Bulgarian regions (B1 to B4) and three North Macedonian regions (SE, South East; E-NE, East-North East; W, West). Black numbers between the circles connect single-locus variants (SLVs) grouped into eight clonal complexes (CC), which are encircled with dashed lines. Gray numbers indicate the number of loci (>1) that differ between connected MLVA haplotypes. Two major genetic clusters, as defined be specific SNPs (GC1 and GC2), are encircled by solid blue (GC1) and red (GC2) lines (see Section 3.5).