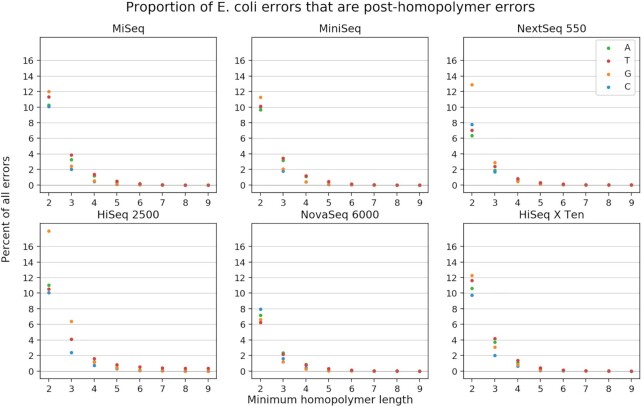
Figure 5.
Frequency of post-homopolymer errors. For each platform, we found all errors where the error base is identical to the preceding genomic base. If the preceding genomic base is part of a single-base repeat (homopolymer), we might call the error a post-homopolymer error, depending on the length of the repeat. This figure shows what percent of all errors are post-homopolymer errors, depending on the threshold one uses for the definition of a post-homopolymer error. Numbers are broken out by error/homopolymer base. For example, if one decides that any error preceded by a run of at least three bases of the same identity qualifies as a post-homopolymer error, then about 2% of all C substitutions in MiniSeq are post-homopolymer errors.