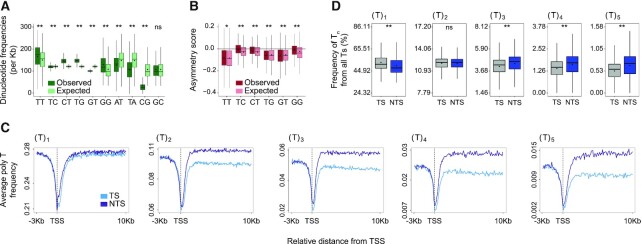
Figure 2.
Polynucleotide frequencies in genes are not explained solely by the frequencies of the single nucleotides composing them. (A) Comparison of the frequency of 10 possible dinucleotides (calculated for both strands, observed, green) compared to the frequency that would be expected by random pairing of the single nucleotides that compose them (expected, mint). (B) Comparison of the observed (burgundy) asymmetry score to the asymmetry score expected (pink) by random pairing of the single nucleotides that compose them. (**P < 0.0001, * P < 0.01 based on a Wilcoxon signed-rank test with Bonferroni correction). (C) Average profiles for polyT sequence frequencies surrounding the TSS of genes for Ts found in the context of T1 and up to T5. (D) The frequency of the Ts found in each of the contexts Tn out of all the Ts on each strand is calculated for T1 to T5. (** P < 0.0001 comparing transcribed and non-transcribed strands based on a paired t-test with Bonferroni correction). In all plots, boxes represent range between 75th and 25th percentile, the line represents the median and the diamond the mean. Outliers were discarded for the presentation.