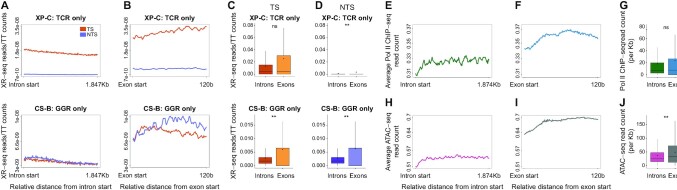
Figure 5.
Differential NER in exons and introns. (A) Average profile of XR-seq after normalization to the underlying TT frequency. Plotted separately are the transcribed strand (TS, pink) and non-transcribed strand (NTS, purple) over the first 1.874 Kb of intron intervals in XP-C (top) and CS-B (bottom) cells. (B) Same as A, except plotted over 120 bases of exons. (C andD) XR-seq normalized to TT frequencies over the transcribed and non-transcribed strands, respectively, of exons and introns in XP-C (top) and CS-B (bottom) cells. (E) Average profile of ChIP-seq read densities of elongating RNA polymerase II (RNAPII) over the first 1.874 Kb of introns. Data plotted for both strands. (F) Same as (E), except plotted over the first 120 bases of exons. (G) Distribution of ChIP-seq reads of elongating RNA polymerase II (RNAPII) over introns and exons. (H) ATAC-seq read densities measuring chromatin accessibility over the first 1.874 Kb of introns. Data plotted for both strands. (I) Same as (H), except plotted over the first 120 bases of exons. (J) Distribution of ATAC-seq reads over introns and exons. In all plots, **P < 0.0001 based on Wilcoxon signed-rank test with Bonferroni correction. boxes represent range between 75th and 25th percentile, the line represents the median and the diamond the mean. Outliers were discarded for the presentation.