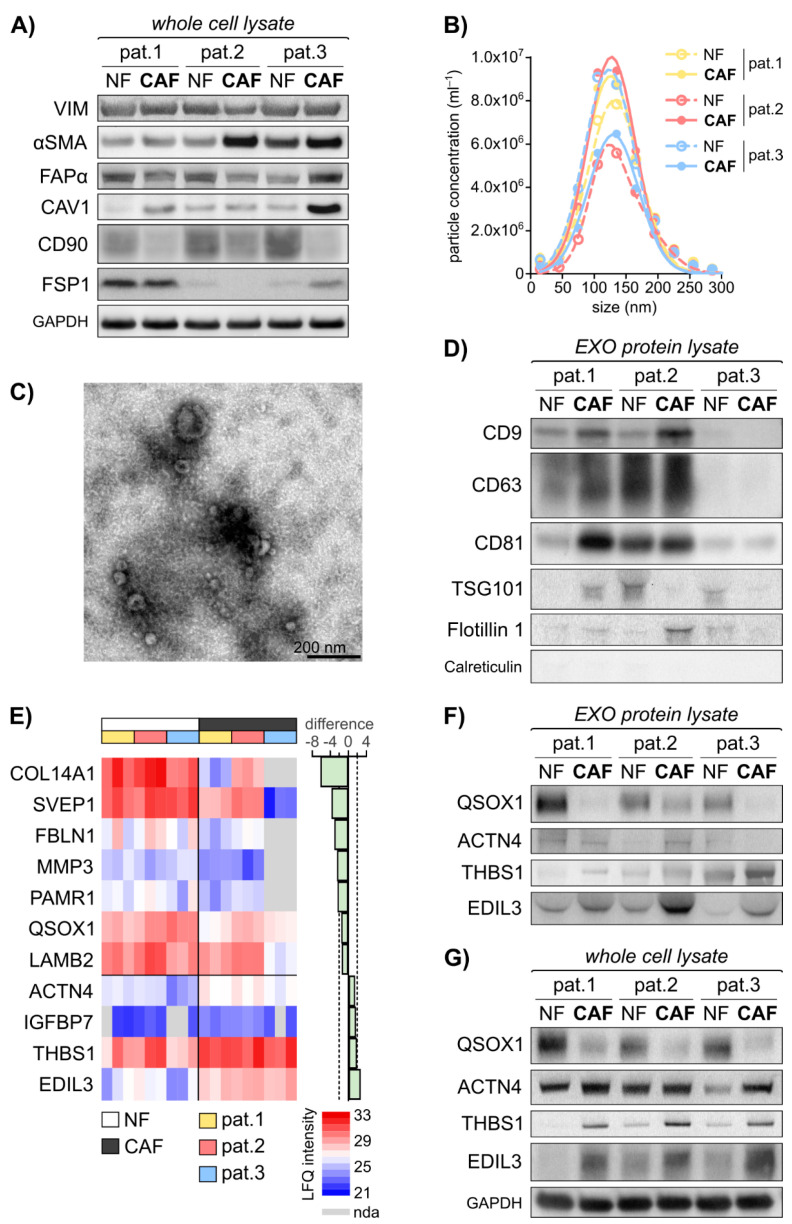
Figure 1.
Primary fibroblast-derived EXOs display an activation status dependent protein cargo. The established fibroblast cell lines were subjected to cellular protein isolation after 48 h incubation of equal cell numbers in normal growth medium. EXOs were isolated using combined differential centrifugation and ultrafiltration approach following 48 h of fibroblast incubation in starvation medium. Exosomal protein was isolated in biological triplicates and subjected to Mass Spectrometry. Statistical Analysis was performed in Perseus. (A) Immunoblot analysis of vimentin (VIM), α-smooth-muscle actin (αSMA), fibroblast activation protein α (FAPα), caveolin 1 (CAV1), cluster of differentiation 90 (CD90)/Thy1 and fibroblast-specific protein 1 (FSP1) in primary fibroblasts, including GAPDH as loading control. (B) Particle size distribution in the isolated EXOs (mean of n = 4–10) as measured by nanoparticle tracking analysis (NTA) using ZetaView®. (C) Representative transmission electron microscopy (TEM) image of fibroblast-derived EXOs. (D) Immunoblot analysis of EXO markers CD9, CD63, CD81, Flotillin 1 and Tumor susceptibility 101 (TSG101), including calreticulin as negative control. (E) Heat map of mass spectrometry data illustrating significantly deregulated vesicular proteins. Proteins with more than 4 undefined values in total or more than 3 undefined values in the NF/CAF subgroups are excluded. Paired t test: q < 0.05, diff. > |1.0|. LFQ: label-free quantification; nda: no data acquired. (F) Immunoblot analysis of quiescin sulfhydryl oxidase 1 (QSOX1), actinin α4 (ACTN4), thrombospondin 1 (THBS1) and EGF-like repeats and discoidin domains 3 (EDIL3) in EXOs. (G) Immunoblot analysis of QSOX1, ACTN4, THBS1 and EDIL3 in primary fibroblasts whole cell lysate, including GAPDH as loading control. The images of uncropped western blot figures are shown in Figure S8.