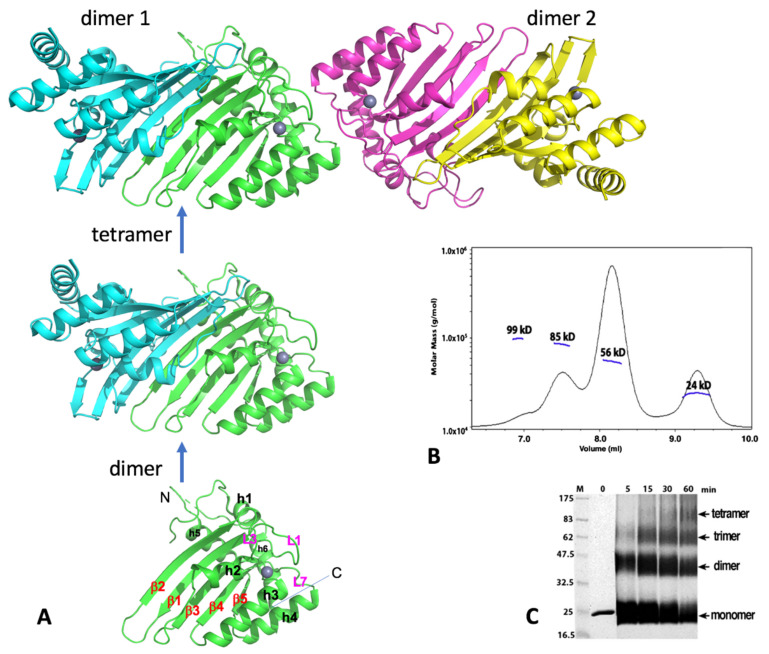
Figure 3.
Human A2 crystal structure. (A) The crystal structure of A2-core tetramer, dimer, and monomer (PDBid 2nyt, residues 40–224). (B) Molecular weights from size-exclusion chromatography coupled with multi-angle light dynamic scattering (SEC-MALS) data for the A2 (full-length, 26 kD for a monomer) at a protein concentration of 1 mg/mL. Four peaks were observed, corresponding to the approximate molecular weight of a monomer (24 kD), dimer (56 kD), trimer (85 kD), and tetramer (95 kD). The dimer species is the predominant form. (C) Time course of glutaraldehyde cross-linking with the A2 protein. Reactions with A2 (2 µg total protein) were performed for the indicated time, with 0.25% glutaraldehyde at room temperature, 25 mM HEPES (pH 7.0), 160 mM NaCl, and 10% glycerol; they were then quenched with 1 M Tris, pH 8.5, with 2X SDS loading buffer and run on a 12% SDS-PAGE gel for 70 min, at 200 V. Proteins were visualized by Coomassie staining. Cross-linking reveals four bands, corresponding to a monomer, dimer, trimer, and tetramer band, respectively. Monomer and dimer appear to be the predominant forms under this cross-linking condition. Both full-length (residues 1–224) and the N-terminal truncated A2-core (residues 40–224) showed similar results. Note: Panels B and C are to-be-published data.