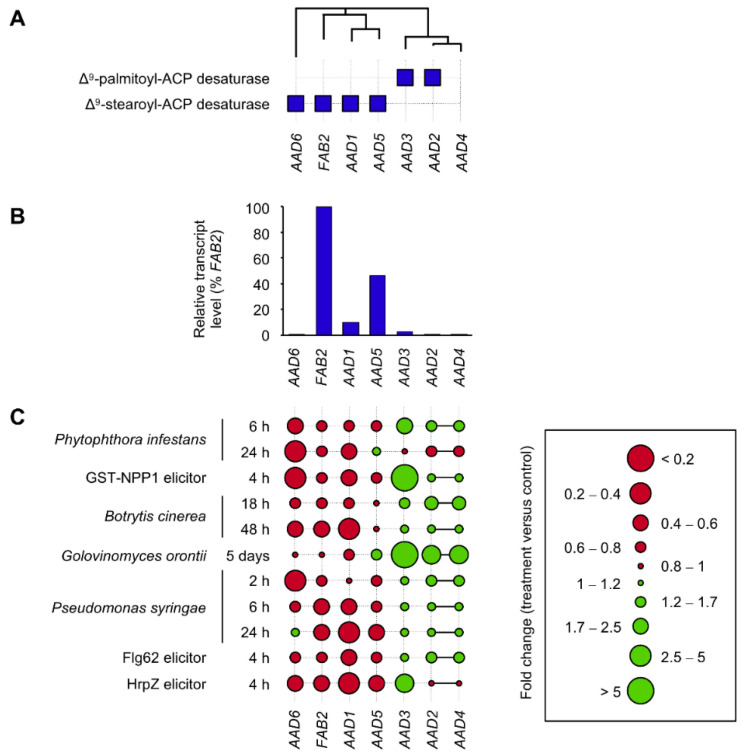
Figure 1.
Transcriptional regulation of AAD genes in the leaves of Arabidopsis thaliana in response to biotic stress. (A) Phylogram, with branch lengths in arbitrary units, using the alignment of the seven A. thaliana AAD sequences (with gaps). Enzymatic activities of the AAD isoforms are indicated below the phylogram (blue squares). The activity of AAD4 remains unknown. (B) Relative AAD transcript levels in control leaves of A. thaliana expressed as a percentage of FAB2 transcript levels. Expression levels were calculated using the data displayed on the Arabidopsis eFP Browser [57]. (C) Variations in AAD transcript levels in leaves of A. thaliana in response to various pathogens and elicitors and expressed in fold changes with respect to control. Bold lines between AAD2 and AAD4 reflect the impossibility of distinguishing between AAD2 and AAD4 transcripts using the Affymetrix technology due to the very similar sequences of these transcripts. Flg62, bacterial derived elicitor; GST-NPP1, oomycete-derived elicitor; HrpZ, bacterial derived elicitor.