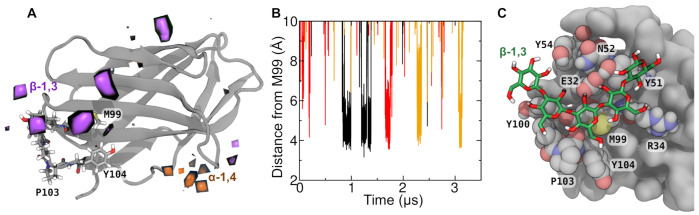
Figure 3.
Binding of β-1,3 oligoglucan to the immune receptor CD28. (A) Regions of high oligoglucan density in unbiased molecular dynamics simulations containing many 5-mer oligoglucan molecules (-1,4 or β-1,3). Regions where the average density of oligoglucan monomers is 8 times their ambient density are highlighted in orange (-1,4) or purple (-1,3). The MYPPPY loop (residues 99–104), which makes up part of the ligand binding site of CD28, is shown explicitly, while the remainder of CD28 is shown in a gray secondary structure representation. (B) Distance between three selected β-1,3 oligomer monomers and residue Met99 during in two replicates of the simulation. We selected curves where there is contact (center-of-mass separation <5 Å) between the glucose moiety and sulfur atom of Met99 continuously for >30 ns. (C) Exemplary simulation frame where a 5-mer β-1,3 oligoglucan is bound to CD28. Residues of CD28 making contact with the oligoglucan are shown as atomic spheres (carbon in gray), while the oligoglucan is shown in a bonds representation (carbon in green).