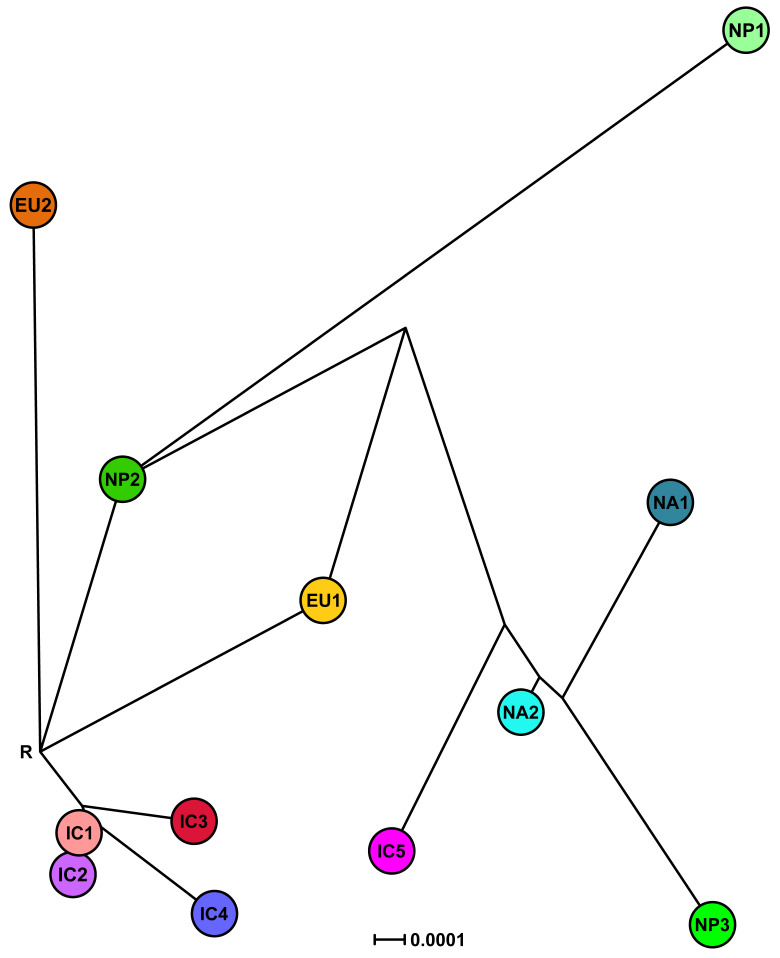
Figure 10.
Split decomposition network (SDN) for the five Indochinese lineages (IC1-5), the three Japanese lineages (NP1-3) and the four known lineages (EU1, EU2, NA1 and NA2) of Phytophthora ramorum based on a seven-locus nuclear (Avh120, Avh121, btub, gwEuk.30.30.1, hsp90, ITS and trp1) dataset. Alternative pathways are shown as parallel edges. The node with the highest number of connections and the most central position was designated as root (R). Missing intermediates represent extant unsampled genotypes, extinct ancestors or “false-positives”. The scale bar represents the split support for the edges.