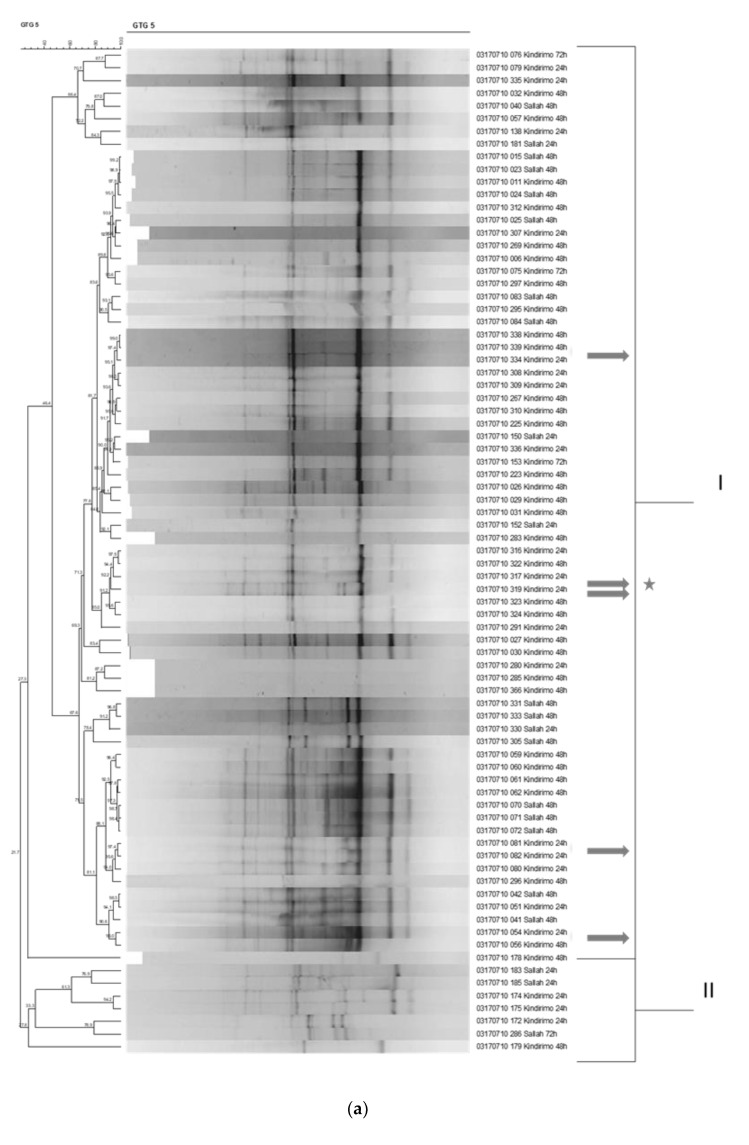
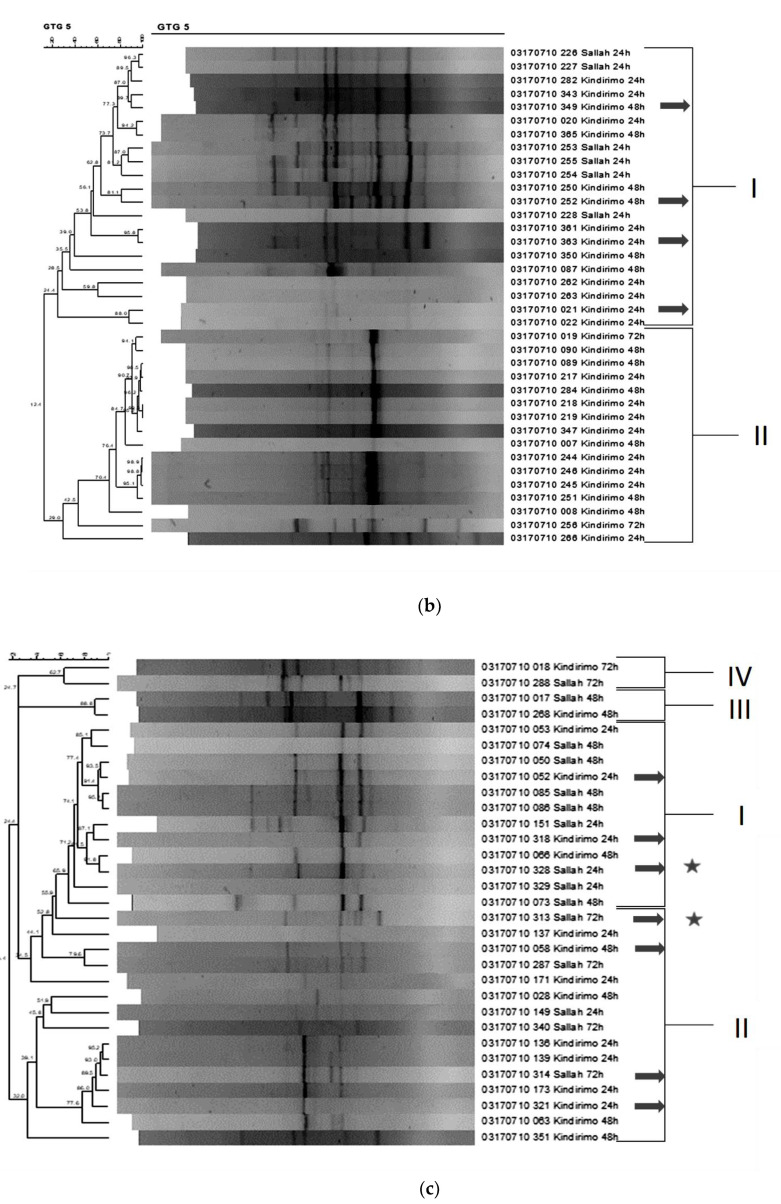
Figure 3.
Dendrogram obtained by unweighted pair-group method using arithmetic averages clustering algorithm (UPGMA) (I-IV clusters) of correlation value r of repetitive element PCR (rep-PCR) fingerprint patterns with primer GTG5 of predominant lactic acid bacterial strains isolated from the fermented milk product nono from Nigeria. (a) Heterofermentative rod-shaped lactic acid bacteria, (b) homofermentative coccus-shaped lactic acid bacteria, (c) homofermentative rod-shaped lactic acid bacteria. Arrows indicate the strains for which the 16S rRNA gene was amplified and sequenced in order to identify. Star indicates strains Limosilactobacillus (L.) fermentum 317, Lactobacillus (Lb.) delbrueckii 328M, and Lb. helveticus 313 that were not selected as (potential) starter strains in this study but were sequenced in a previous study to confirm 16S rRNA gene sequencing results and to establish genome sequencing methods [23].