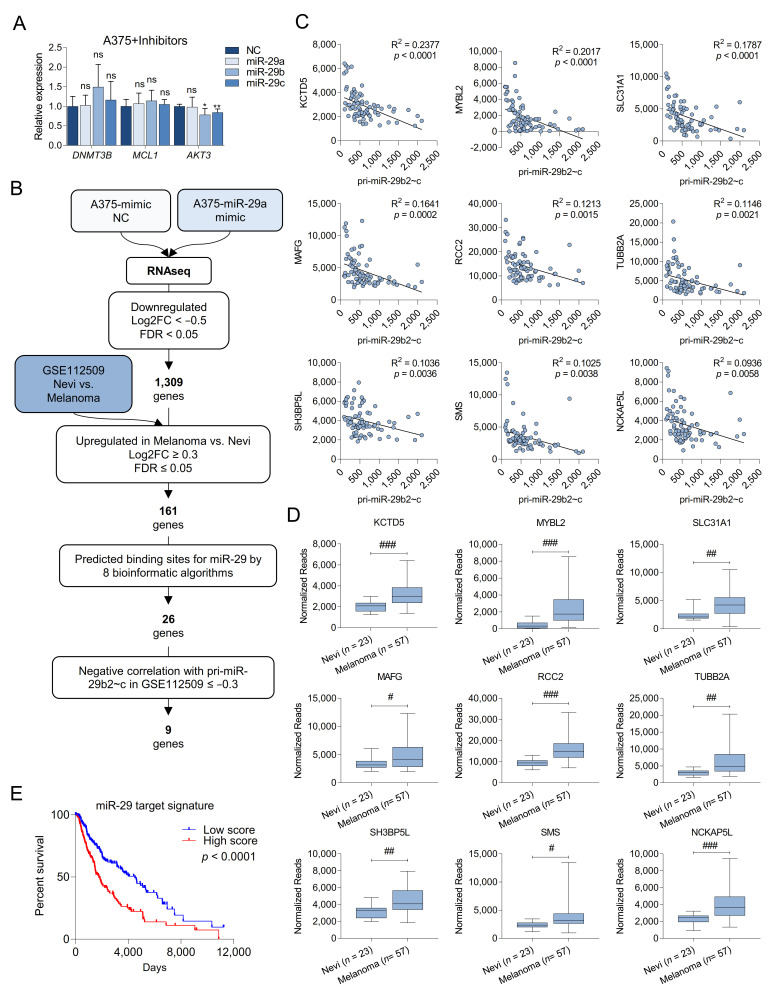
Figure 3.
Identification of new miR-29 targets in melanoma. (A) Quantification by qRT-PCR of three validated miR-29 target genes of miR-29 after overexpression of miR-29 mimics in A375 cells. Cells overexpressing a negative control mimic were used as control; (B) selection flowchart to identify target genes of miR-29 involved in melanoma progression; (C) correlation between the normalized reads of pri-miR-29b2~c and the normalized reads of the top nine identified putative miR-29 targets in the GSE112509 dataset; (D) expression of the top nine identified putative miR-29 targets in 23 nevi and 57 melanomas obtained from the GSE112509 dataset; (E) survival analysis from TCGA (PanCancer Atlas, n = 363) comparing melanoma patients with high or low expression of the nine putative miR-29 targets. ns, not significant; * p < 0.05; ** p < 0.01; FDR, false discovery rate; # FDR < 0.05; ## FDR < 0.01; ### FDR < 0.001.