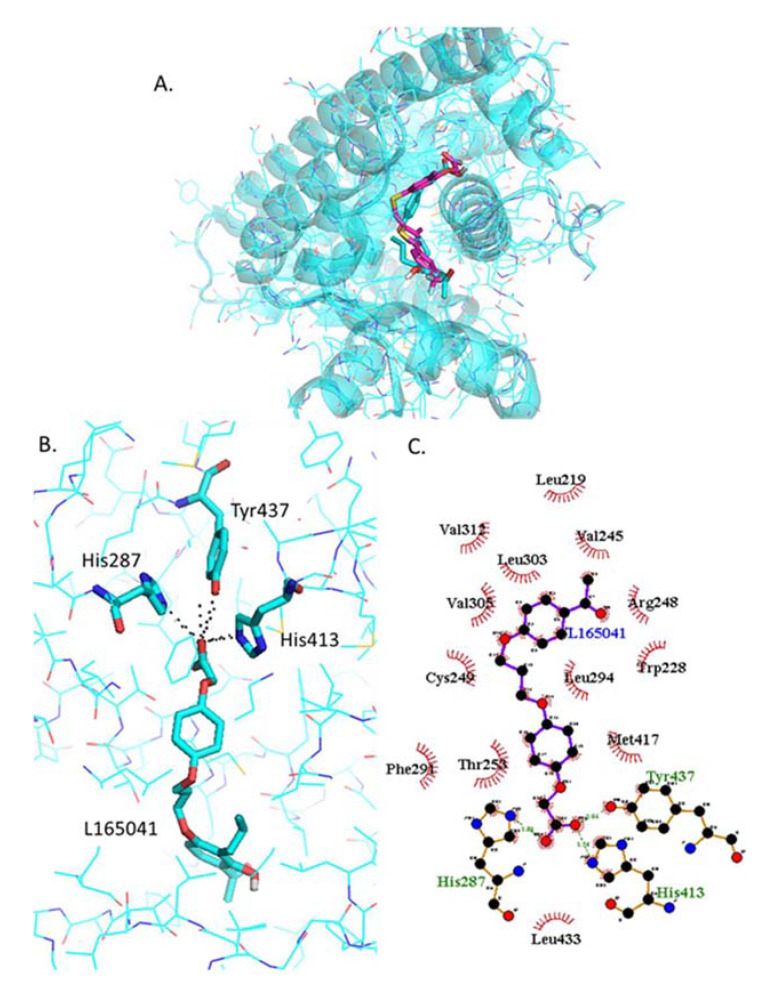
Figure 5.
Analysis of L−165042 docked into PPARβ/δ (PBD:3TKM). (A) Representation of the most stable L−165041 docking conformation (cyan sticks) compared to the GW0742 of the crystal structure (pink). (B) 3D detail of the amino acids forming polar bindings with L−165041 calculated by Pymol. Color coding of atoms: red O, Blue N, mustard S, white F, pink C of GW0742 from the crystal structure, cyan sticks C of L−165041 docked into the crystal structure, cyan lines C from PPARβ/δ. (C) Schematic 2D representation of the interaction between PPARβ/δ LBD and -L−165041 calculated using Ligplot+. The green dashed lines indicate polar interactions and the red spoked arcs indicate hydrophobic interactions. Colour coding of atoms: red O, blue N, yellow S, green F, black C.