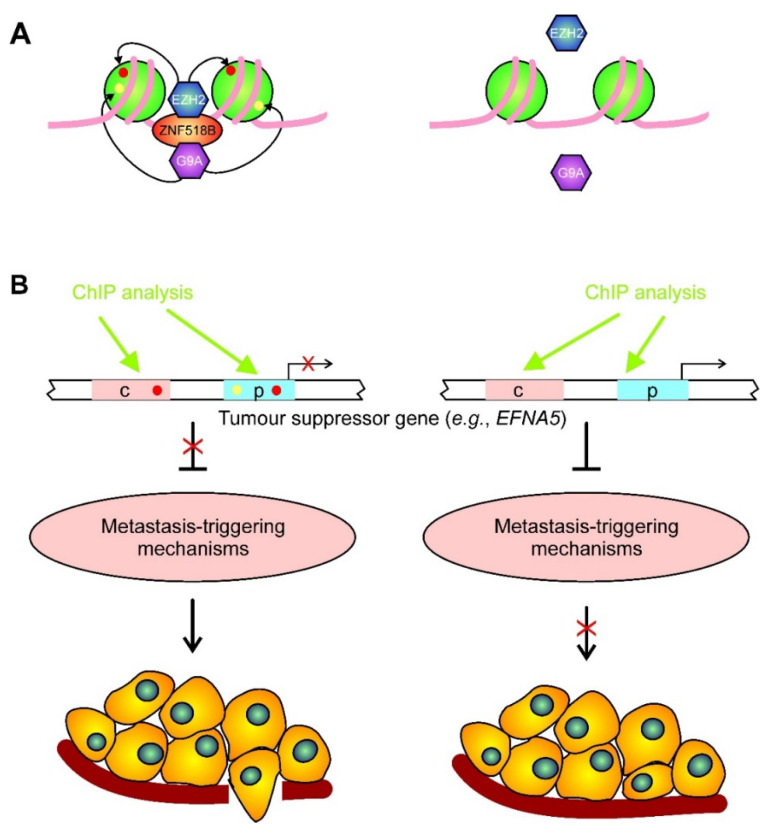
Figure 6.
A possible mechanism for ZNF518B oncogenic role. (A) Schematic drawing of the starting hypothesis for the chromatin immunoprecipitation (ChIP) experiments. ZNF518B binds a specific DNA target in a tumour suppressor gene and recruits a histone methyltransferase (EZH2 or G9A), which catalyses the introduction of the repressive marks (H3K27me3, red ovals, or H3K9me3, yellow ovals, respectively) in the histones of the neighbouring nucleosomes (left). In the absence of ZNF518B, the histone methyltransferases are no longer recruited, and the repressive marks are not introduced (right). (B) The presence or absence of the methyltransferases and/or of the repressive epigenetic marks, either in the promoter (p) or in some control region (c), determines the expression of a suppressor gene. These epigenetic features can be experimentally detected by ChIP analysis. The expression of the suppressor gene, in turn, controls the triggering of mechanisms that eventually result in the dissemination and metastasis of tumour cells as a consequence of ZNF518B activity.