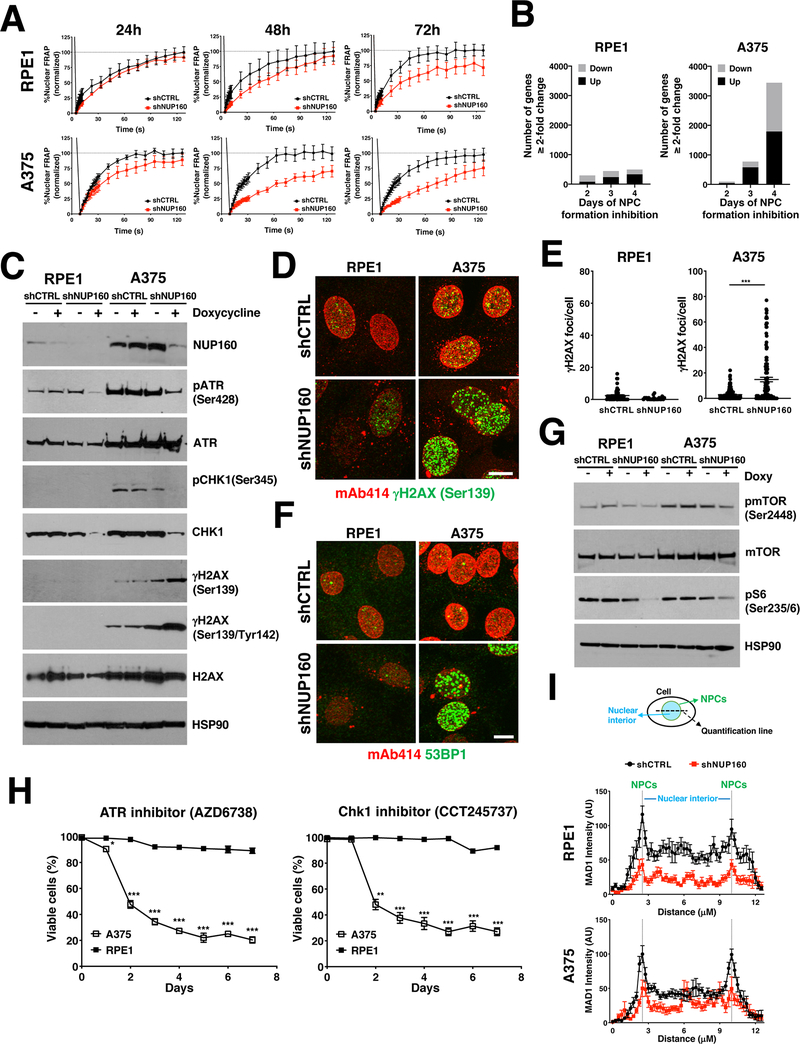
Figure 6. Reduction in NPC number results in multiple cellular alterations.
A, Fluorescence recovery after photobleaching (FRAP) analysis showing nuclear import of the NES-Tomato-NLS reporter in RPE1 and A375 cells after treatment for the indicated number of days with Control or NUP160 siRNAs. The data are expressed as percentage of FRAP relative to maximum recovery of Control (n = 8–16 cells). B, RNAseq analysis of genes significantly changed ≥ 2-fold (q-value < 0.05) in RPE1 and A375 cells treated with NUP160 shRNAs for the indicated number of days compared to Control cells. C, Western blot analysis of DNA damage response proteins in RPE1 and A375 cells untreated or treated with doxycycline for 72 hours to induce Control or NUP160 shRNAs. D, Immunofluorescence analysis of NPCs (mAb414) and γH2AX (Ser139) in RPE1 and A375 cells after 96 hours days of treatment with Control or NUP160 shRNAs. Images show the maximum projection of entire nuclei. Scale bar, 10 μm. E, Quantification of images from (D) (n = 59–79 cells). F, Immunofluorescence analysis of NPCs (mAb414) and 53BP1 in RPE1 and A375 cells after 96 hours days of treatment with Control or NUP160 shRNAs. Representative images show the maximum projection of entire nuclei. Scale bar, 10 μm. G, Western blot analysis of mTOR signaling proteins in RPE1 and A375 cells untreated or treated with doxycycline for 3 days to induce Control or NUP160 shRNA. H, Viability was determined by automated counting of Trypan blue inclusion/exclusion of proliferating A375 and RPE1 cells grown with 10 μM of ATR or CHK1 inhibitors for the indicated time points. I, Top panel shows schematic illustration of method used to quantify MAD1 association with nuclear pores. Bottom panel shows quantification of MAD1 immunofluorescent images of RPE1 and A375 cells (n = 12–14 cells) 72 hours after induction of Control or NUP160 shRNAs with doxycycline. Dashed lines indicate edge of nuclear periphery/NPCs. Data are mean ± s.e.m (A, E) or mean ± s.d. (H, I). * P ≤ 0.05, ** P ≤ 0.01, *** P ≤ 0.001 by unpaired Student’s t test (E) or multiple unpaired Student’s t tests with Holm-Sidak method to correct for multiple comparisons (H). Unless otherwise stated, experiments are representative of a minimum of 3 independent repeats.