Figure 1.
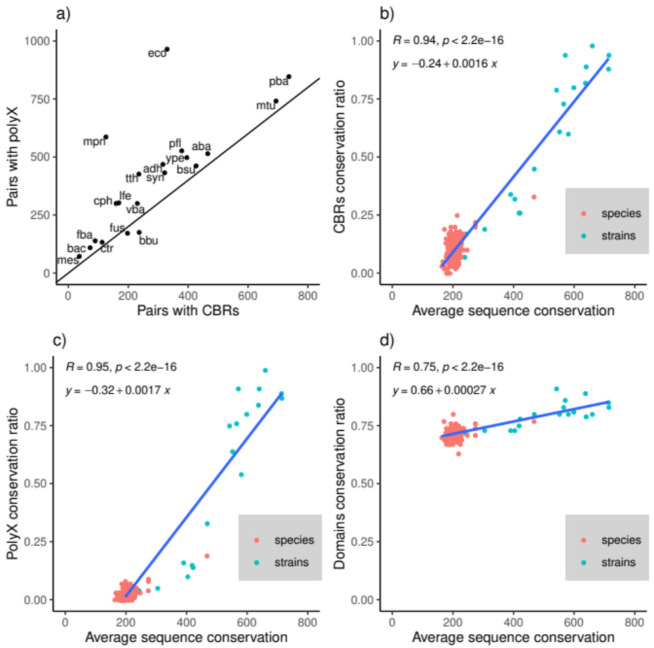
(a) Number of orthologous pairs between strains with polyX versus the number of pairs with compositionally biased regions (CBRs); in black, line x = y; (b) CBRs; (c) polyX, and (d) domain conservation ratio versus average full-sequence conservation in orthologs from pairs of strains (or species). Sequence conservation is measured as the average bit score of all orthologous pairs. The regression line is plotted in blue.
