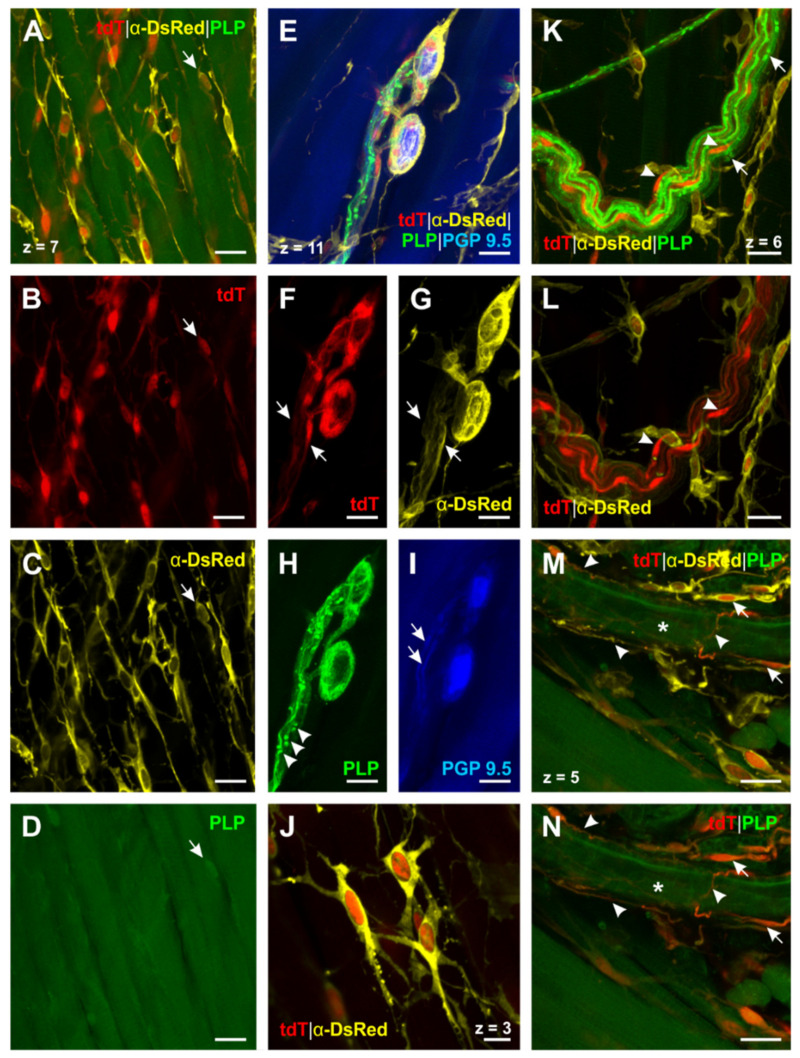
Figure 4.
PLP+ -glial cells in the esophagus identified by triple staining for DsRed, PLP and PGP 9.5 in PLP-CreERT2 x tdT mice (N° ②, Table 2). (A)–(D) (PGP 9.5 not shown): PLP+ -glial cells form a meshwork throughout the esophagus. Some of these cells, located in the tunica muscularis, are arranged in parallel with the muscle fibers. While only a few of these cells can be detected by PLP antibody staining ((A–D), short arrow), transgenic PLP-CreERT2 x tdT mice reveal their distribution (cf. A–C vs. D). (E–I): PLP+ -glial cells interact very closely with enteric neurons as the processes of the EGCs are woven around the neurons (E–H). Interconnecting strands of enteric neurons (I, short arrows) are accompanied by PLP+ -glial cells (F and G, short arrows). PLP antibody staining indicates the contact zone of these cells (H, arrowheads). (J) (PGP 9.5 and PLP not shown): Close-up of PLP+ -EGCs; these cells are interconnected by their fine, filiform processes and therefore form a network of glial cells. (K–L) (PGP 9.5 not shown): PLP+ -myelin sheaths of a vagal nerve fiber bundle in the tunica adventitia can be identified by the PLP antibody (K, nodes of Ranvier are marked by short arrows). TdT expression shows the cell bodies of the peripheral myelinating Schwann cells ((K and L), arrowheads). (M and N) (PGP 9.5 not shown): Blood vessel-connected EGCs have a delicate morphology ((M and N), short arrows) and very long filiform processes, which appear woven around the outer vessel wall ((M and N), arrowheads). The lumen of the blood vessel is marked by the asterisk. DsRed: Discosoma sp. red fluorescent protein; PGP 9.5: protein gene product 9.5; PLP: proteolipid protein; tdT: tdTomato. Z-step = 1 µm; scale bars 20 µm (A–I, K–N), 10 µm (J).