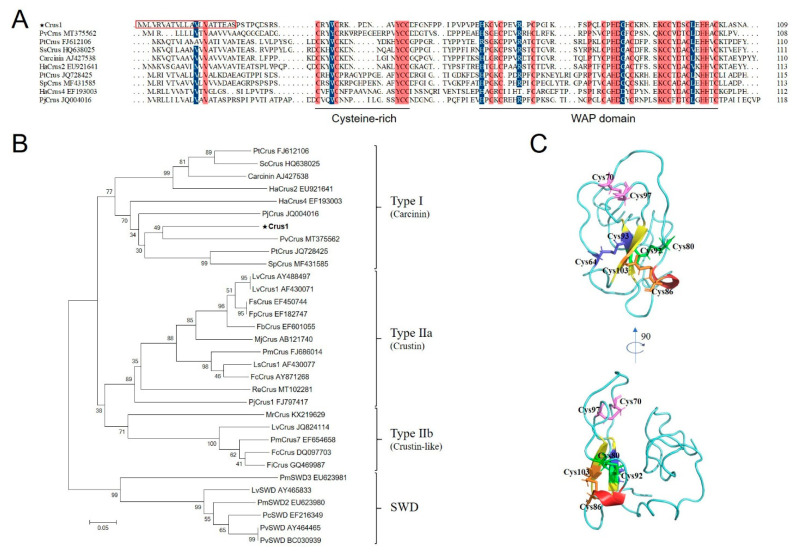
Figure 1.
Sequence, phylogenetic, and structural analysis of Crus1. (A) Alignment of Crus1 with Type I crustins. Dots denote gaps introduced for maximum matching. The consensus residues are shaded red, the residues that are ≥75% identical among the aligned sequences are shaded blue. The signal peptide sequence of Crus1 is boxed with red lines. (B) Phylogenetic analysis of Crus1 homologues. The phylogenetic tree was constructed with MEGA 6.0 using the neighbor-joining method. Numbers beside the internal branches indicate bootstrap values based on 1000 replications. The GenBank accession numbers of the crustins used in (A,B) are indicated after the names of the crustins. (C) The predicted structure of Crus1 was built using I-TASSER. The disulfide bonds in the WAP domain are shown in blue (C64–C93), pink (C70–C97), green (C80–C92) and orange (C86–C103).