Figure 3.
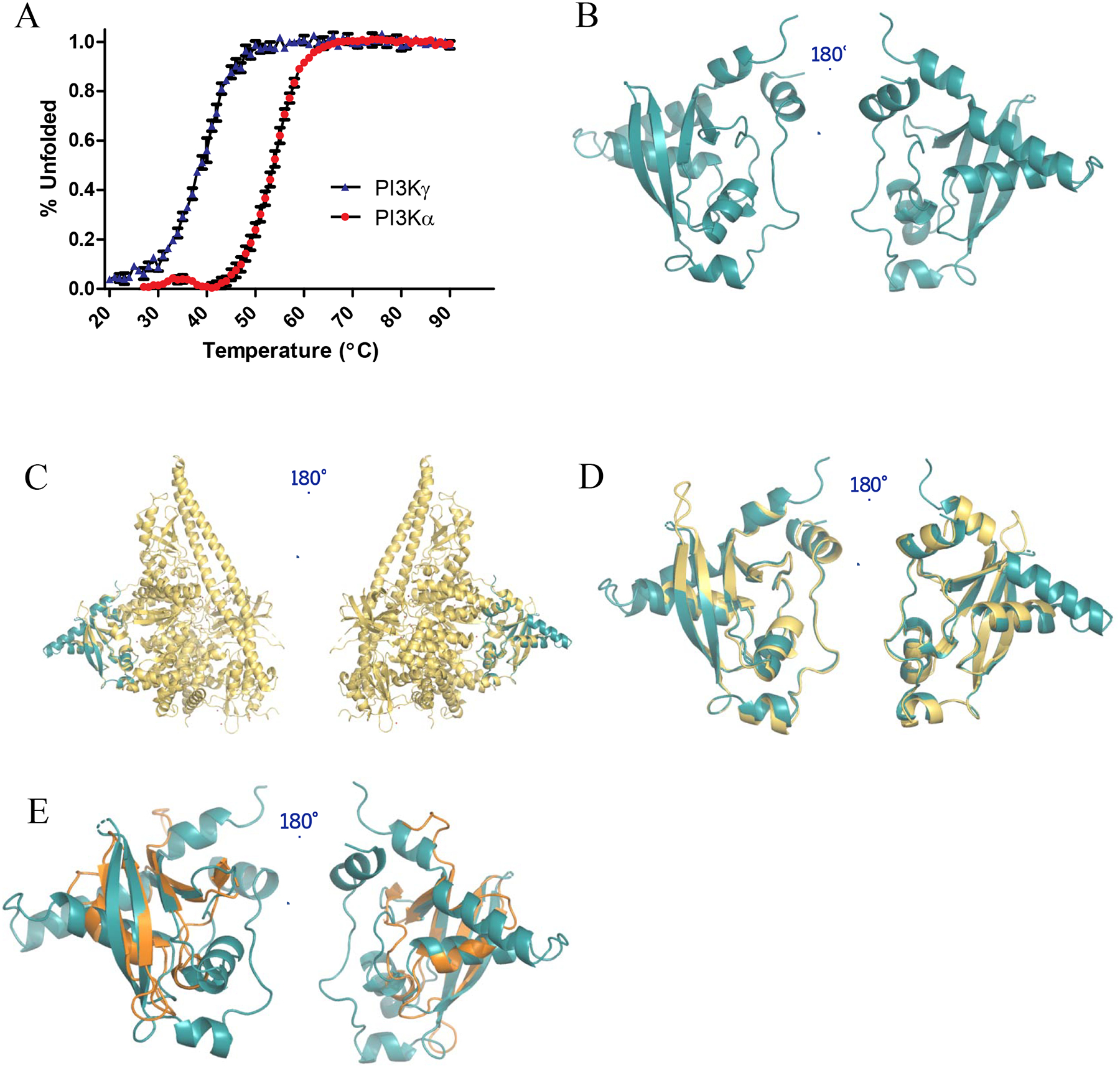
Structural characterization of PI3Kα-RBD. (A) CD thermal unfolding as quantified by the loss of mean residue ellipticity at 222 nm for 30 μM PI3Kα-RBD (red) and PI3Kγ-RBD (blue) over a temperature range of 20–90°C at pH 7.4. Data was normalized to the maximum and minimum ellipticity. Thermal denaturation curves and the midpoint of the thermal transition (Tm) were calculated by fitting the data using the Gibbs-Helmhotz equation. Results were graphed in GraphPad Prism and reported as the mean ± S.E. (error bars) (N=3 for PI3Kα, N=2 for PI3Kγ, each performed in duplicate). (B) X-ray crystallographic structure of the PI3Kα-RBD (residues 157–300) (PDB: 6VO7). The structure was refined to a resolution of 2.31 Å, with residual values of 19.65 % (Rwork) and 25.52 % (Rfree). The structure is complete, save for 4 unobservable residues in the flexible loop region between β-strands 1 and 2, and 16 unobservable residue sidechains. Superposition of (C) PI3Kα-RBD crystal structure (teal) onto the full length PI3Kα structure (PDB: 5XGH, yellow), (D) PI3Kα-RBD crystal structure (teal) overlayed with the RBD contained within the PI3Kα full-length crystal structure (PDB: 5XGH, yellow, correlated construct residues 157–300, Cα least squares quadratic (LSQ) RMSD 1.22 Å), (E) PI3Kα-RBD crystal structure (teal) overlayed with B-RAF-RBD (PDB: 3NY5, gold). The 4 β-strands align well between the B-RAF-RBD and PI3Kα-RBD, with similar orientations observed for the conserved helices adjacent to the beta sheets.
