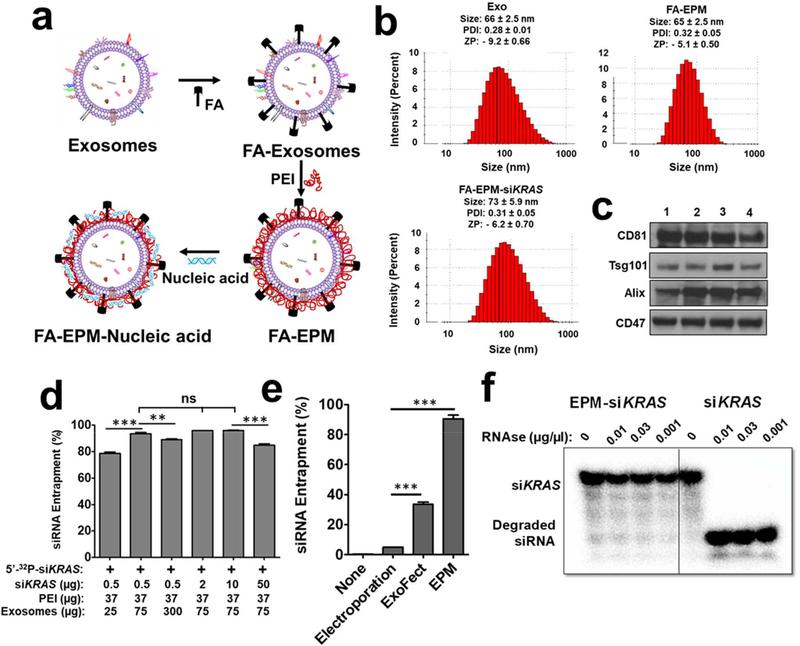
Figure 1.
(a) Graphical representation of folic acid (FA) covalently attached to the exosome-polyethyleneimine matrix (EPM) as a transfection vector for delivery of nucleic acid. (b) Size, polydispersity index (PDI), and zeta potential (ZP) of exosomes, FA-EPM, and FA-EPM-siKRAS, analyzed by Zetasizer. Data represent mean ± SD from 3 preparations. (c) Surface-bound hallmark protein markers in bovine colostrum exosomes, as analyzed by western blot; exosomes isolated from different batches (1 – 4) of bovine colostrum powder were analyzed. (d) The entrapment efficiency of EPM with siKRAS; 5’−32P-labeled siKRAS was included as a tracer. Entrapment efficiency was calculated by the distribution of the radioactivity in the EPM-siKRAS recovered by precipitation with ExoQuick™ and in the supernatant, followed by radioactivity quantitation by Packard InstantImager. (e) The relative entrapment efficiency of the EPM versus the conventional methods involving electroporation and Exo-Fect™. Test siRNA was entrapped by EPM, electroporation, and Exo-Fect™; radiolabeled siRNA was included as a tracer. Entrapment efficiency was determined as described in Fig. 1d legend. (f) Susceptibility of 5’−32P-labeled siKRAS entrapped in EPM to enzymatic degradation. Following a brief incubation of EPM with the radiolabeled siKRAS, the reaction mixture was incubated with indicated concentrations of RNase A for 30 min. The reaction products were resolved by polyacrylamide gel electrophoresis after adding heparin to dissociate the siKRAS from PEI, and radioactive products were detected by Packard InstantImager. Statistical analysis was performed by one-way ANOVA followed by Tukey’s multiple comparison test (d) and using student’s t-test (e) to compare between the indicated groups. **, p < 0.01; *** p < 0.001.