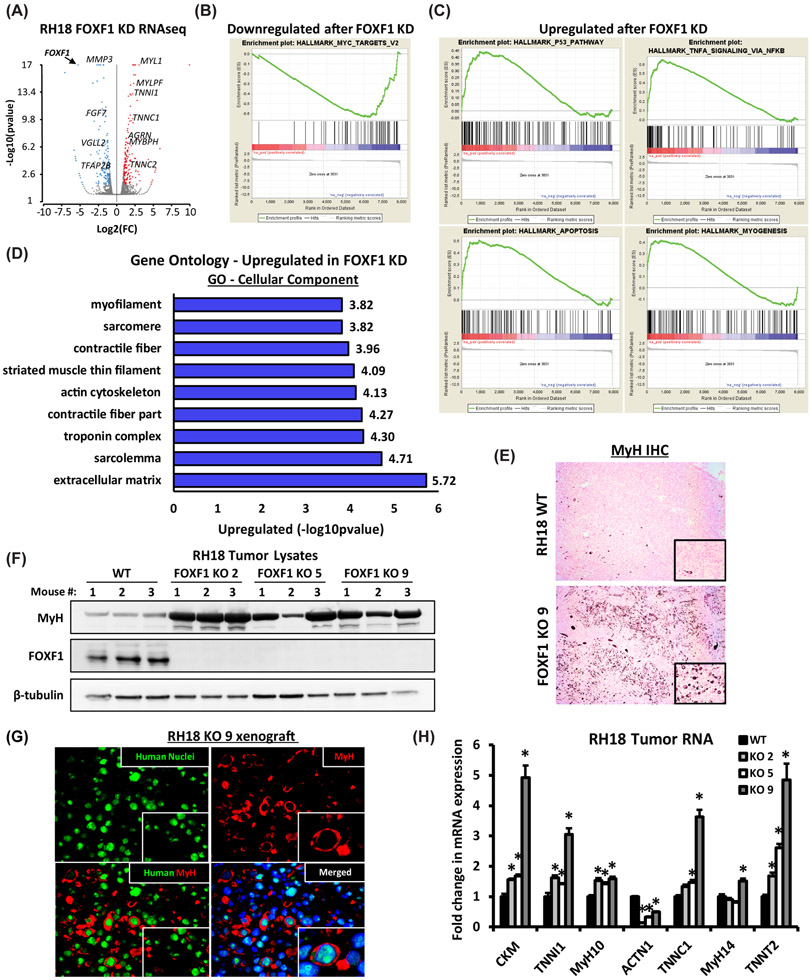
Figure 4. FOXF1 regulates expression of genes critical for FP-RMS tumor growth and metastasis.
(A) Volcano plot shows differentially expressed genes after FOXF1 knockdown in RH18 cells. (B) Gene Set Enrichment Analysis (GSEA) analysis shows downregulation of MYC gene network after FOXF1 knockdown. (C) GSEA analysis shows upregulation of P53 pathway, apoptosis and myogenesis after FOXF1 knockdown. (D) Gene ontology analysis of genes significantly upregulated after FOXF1 knockdown. (FC>1.5; p<0.05). (E) Loss of FOXF1 induced spontaneous differentiation of FP-RMS tumor cells in vivo. Deletion of FOXF1 (FOXF1 KO, clone 9) increased number of myosin heavy chain (MyH)-positive cells in orthotopic FP-RMS tumors compared to control RH18 WT cells. (F) Western blots show increased levels of MyH protein in micro-dissected FOXF1-KO tumors generated from three different clones (KO 2, KO 5 and K9) (n=3 mice per group). β-tubulin is used as a loading control. (G) MyH-positive cells are human in origin as shown by co-staining with a pan-human antibody using immunofluorescent staining of FP-RMS xenografts. (H) Deletion of FOXF1 increased mRNAs of mature skeletal muscle genes. qRT-PCR was performed using RNA isolated from micro-dissected FP-RMS tumors. Values normalized to β-actin. (n=4 mice per group). Data reported as mean ± SEM. * p<0.05.