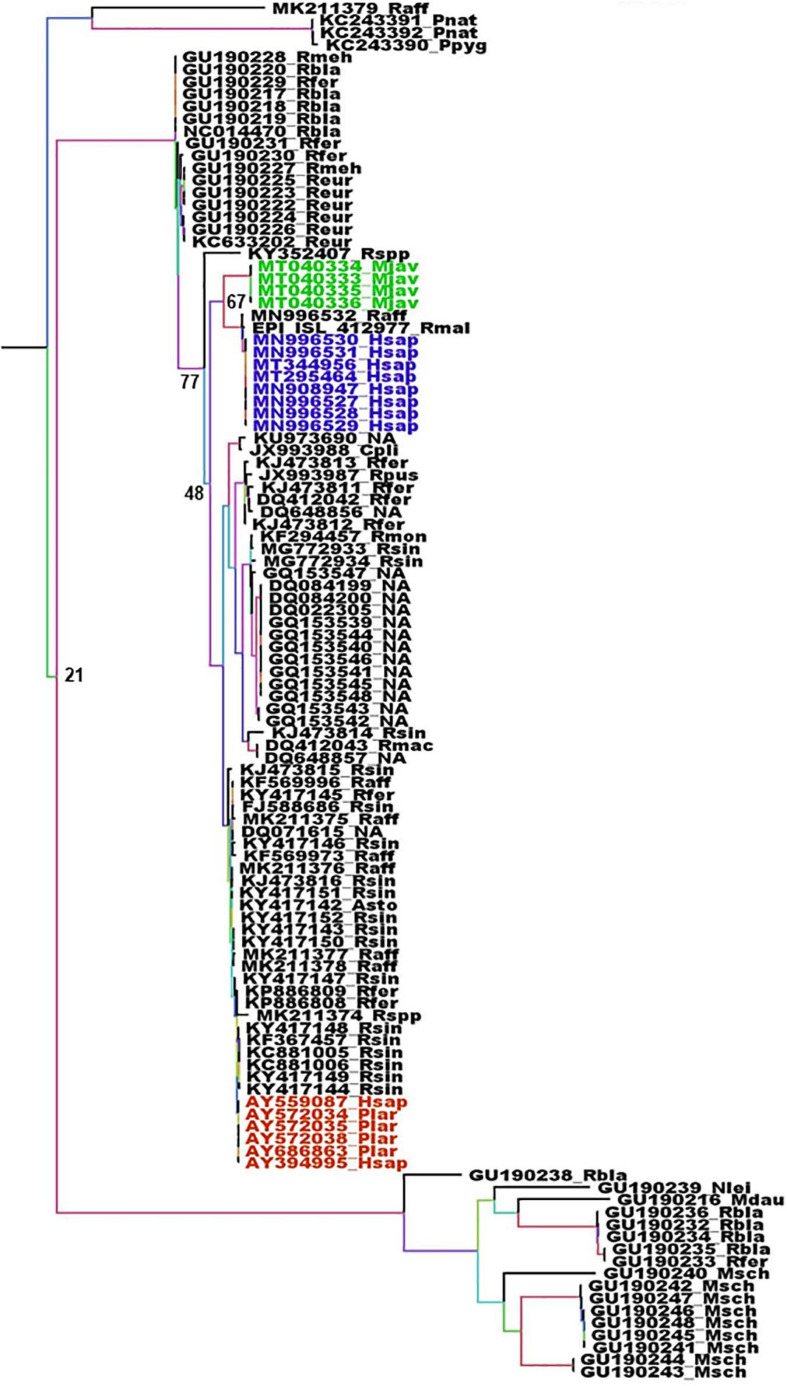
FIGURE 3.
Phylogenetic analysis of Sarbecoviruses RdRp genes. The alignment of the full RdRp genes was performed with MUSCLE from the SeaView package (Gouy et al., 2010). The tree was built using the maximum likelihood method under the GTR model with 500 repeats. The tree was rooted using the RdRp sequence of a MERS-CoV from Camelus dromedarius (KT368883) as outgroup. Violet: RdRp sequences from human SARS-CoV-2. Green: RdRp sequences from pangolins’ Sarbecoviruses. Red: RdRp sequences from SARS-CoV. Sample names are built with the GenBank accession number followed by a four-letter code identifying the species. The species codes are as follows: Asto, Aselliscus stoliczkanus; Cpli, Chaerephon plicata; Hsap, Homo sapiens; Mdau, Myotis daubentonii; Mjav, Manis javanica; Msch, Miniopterus schreibersii; Nlei, Nyctalus leisleri; Plar, Paguma larvata; Pnat, Pipistrellus nathusii; Ppyg, Pipistrellus pygmaeus; Raff, Rhinolophus affinis; Rbla, Rhinolophus blasii; Reur, Rhinolophus euryale; Rfer, Rhinolophus ferrumequinum; Rmac, Rhinophilus maculatus; Rmal, Rhinolophus malayanus; Rmeh, Rhinolophus mehelyi; Rmon, Rhinophilus monoceros; Rpus, Rhinolophus pusillus; Rsin, Rhinolophus sinensis; Rspp, Rhinolophus unidentified species; NA, Not available.