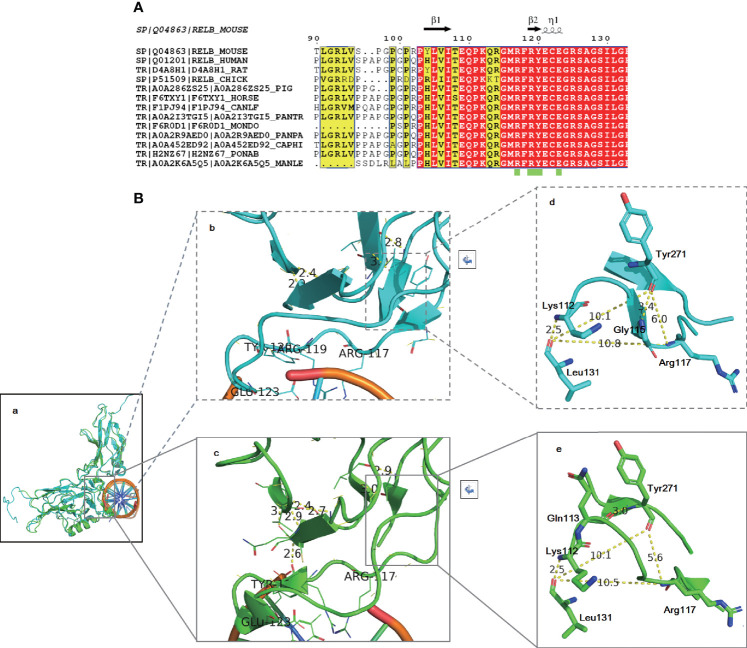
Figure 5.
Amino acid alignment (A) and structural modeling (B) demonstrated that the identified variant may be pathogenic. Amino acid alignment showed that the region of RelB around the identified variant is highly conserved among species (A). The RelB structure is highly conservative (Ba). The local secondary structures around the DNA binding region are slightly different: R114-M116 form a β sheet in 3do7 (Bb) and C122-G124 make up an α helix in 2v2t (Bc). However, the trend of amino acids is completely consistent. The distances between K117, L131, and Y271 are almost the same in two different states (Bd, Be).