Abstract
Recently, numerous evidence has revealed that excessive reactive oxygen species (ROS) production and mitochondrial disruption during acute lung injury (ALI) and its most severe form, acute respiratory distress syndrome (ARDS) will aggravate the inflammatory process. To identify whether antioxidation can be one of the treatment strategies during this progress, we chose mitoQ, a mitochondria-targeted antioxidant that was proved to be effective in reducing ROS generated in mitochondria, as a ROS scavenger to investigate the role of antioxidation in ALI. We demonstrated that overoxidation occurred during the process of ALI, which could be reduced by mitoQ. In the meantime, apoptosis of endothelial cells of ALI mice, accompanied by hyperpermeability of pulmonary vascular and impaired pulmonary function, was partially reversed following an intraperitoneal injection of mitoQ. Moreover, in in vitro study, lipopolysaccharides (LPS) induced excessive ROS production, mitochondrial dysfunction and apoptosis in human pulmonary microvascular endothelial cells (HPMECs), which were rectified by mitoQ. To explore underlying mechanisms, we proceeded RNA-sequencing and found significantly upregulated expression of musculoaponeurotic fibrosarcoma F (MafF) in mitoQ treated group. Additionally, mitoQ inhibited the degradation and increased nuclear translocation of NF-E2-related factor 2 (Nrf2) and upregulated its downstream antioxidant response elements (AREs), such as heme oxygenase (HO)-1 and NAD(P)H:quinone oxidoreductase (NQO)-1. This effect was abolished by transfecting HPMECs with Nrf2 or Maff siRNA. In Nrf2 deficient mice, the protective effects of mitoQ on LPS model of ALI were largely vanished. Taken together, these results provide insights into how antioxidation exerts beneficial effects on ALI via maintaining mitochondrial hemostasis, inhibiting endothelial cells apoptosis, attenuating the endothelial disruption and regulating lung inflammation via Nrf2-MafF/ARE pathway.
Keywords: MitoQ, ALI, Endothelial cells, Apoptosis, Nrf2
1. Introduction
Acute lung injury/acute respiratory distress syndrome (ALI/ARDS) is recognized as a syndrome of acute hypoxemic respiratory failure which features diffuse pulmonary infiltrates, increased capillary permeability, impaired gas exchange and lung inflammation [1]. The mortality of ALI/ARDS is determined by the severity of lung injury, the extent of nonpulmonary organ dysfunction, preexisting medical conditions, and the quality of supportive care [2]. According to randomized trials till now, no effective therapies for ALI/ARDS have yet been identified, except for optimize mechanical ventilation and fluid therapy, which have been proved to result in improved clinical outcomes [3,4]. Because of the limited treatment strategies and high severity, mortality of ALI/ARDS remains high at 30–40% [2]. Thus, more effective treatments are in urgent need.
Although the molecular mechanisms and pathophysiology process related to ALI/ARDS are not completely understood, there is accumulating evidence implicating a role for mitochondrial oxidative damage in ALI [5]. Reactive oxygen species (ROS) refer to free radicals that contain one or more unpaired electrons, including hydroxyl radicals (·OH), singlet oxygen (O2·), and hydrogen peroxide (H2O2) [6]. Studies using animal models of ALI indicate that pharmacological agents either block ROS production or scavenge oxidants ameliorate or prevent ALI [7,8]. During ALI, when critical tissue injury and exacerbation of inflammation occur, cell endogenous antioxidant system is overwhelmed by the overproduced ROS generated by various kinds of cells including activated neutrophils, macrophages, eosinophils, alveolar epithelial and endothelial cells, etc. and results in cell death and lung dysfunction [9]. Excessive ROS can cause oxidation and cross-linking of proteins, lipids, DNA, and carbohydrates, which triggers disruption of normal molecular structure and cellular physiological process, such as increased membrane rigidity, altered permeability, and dysfunction of receptor and enzymes [10,11]. In consequence, targeting endogenous ROS might represent a potential therapeutic strategy in stabilizing the microenvironment in the lung during ALI/ARDS.
Mitochondria are the major resource of ROS, and mitoQ is a mitochondria targeted antioxidant composed of lipophilic triphenylphosphonium cation which can easily move through phospholipid bilayers driven by mitochondrial membrane potential and an ubiquinol antioxidant [12]. Once accumulating within mitochondria, mitoQ will be adsorbed to the surface of the inner membrane where it will be continually transformed to the antioxidant quinol form in the respiratory chain by complex II [13], which may react directly with superoxide (O2·) [14]. It is also shown that mitoQ is able to protect cells against peroxynitrite damage but it can hardly eliminate hydrogen peroxide (H2O2) like other quinols [15]. Growing evidence shows that mitoQ exerts beneficial effects on a wide range of diseases including Parkinson's disease [16], diabetes [17,18], and cardiac ischemia-reperfusion injury [19] where mitochondrial oxidative damage plays a crucial role in the pathology. In addition, in some inflammatory diseases, such as sepsis, mitoQ is also proved to be effective both in vitro and in vivo [20]. Nrf2 is retained in the cytosol bounding to Kelch-like ECH-associated protein 1 (Keap1), which represses activity of Nrf2 by promoting the proteasomal degradation of Nrf2 [21]. In the presence of ROS, Nrf2 will dissociate from Keap1, translocate to the nucleus, dimerize with a small protein Maf, namely MafF, MafG and MafK, and initiate transcription of many cytoprotective genes at antioxidant response elements (AREs) loci, including glutathione peroxidase (GPX), GSH-S-transferase, heme oxygenase (HO)-1, NADPH quinine oxidoreductases (NQO)-1, etc. [22,23]. Previously it was reported that mitoQ improved neurological deficits, attenuated brain edema and decreased cortical neuronal apoptosis through activating the Nrf2/ARE pathway [24]. But the underlying molecular mechanism of mitoQ in ALI model is unclear. Therefore, in this study, we explored the effect of mitoQ on ALI as well as the mechanisms how mitoQ exerts its function.
2. Materials and methods
2.1. Animal experimental design
The animal experiments were approved by the Ethics Committee of the Second Affiliated Hospital, Zhejiang University School of Medicine, and all the experimental processes were carried out according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. Pathogen free 6–8 weeks old C57BL/6 female mice bought from the Animal Center of Slaccas (Shanghai, China) and Nrf2−/− mice obtained from the Jackson laboratory were used for the animal experiments. They were divided into four groups: a PBS/PBS group, a PBS/mitoQ group, an LPS/PBS group and an LPS/mitoQ group. Mice were injected intraperitoneally (i.p.) with or without mitoQ (5 mg/kg, Medchemexpress, NJ, USA) 30 min earlier, and then anesthetized by 1.5% pentobarbital sodium (Sigma-Aldrich, USA), and LPS (from Escherichia coli 0111: B4; Sigma-Aldrich) diluted in sterile 1x PBS was instilled intratracheal (i.t.). Twelve hours after LPS (5 mg/kg) administration, mice were sacrificed for further analysis. For survival study, mice were injected intratracheally with a lethal dose of LPS (50 mg/kg). The survival rate was recorded every 24 h until 7 days post-LPS injection.
2.2. Histological analysis
For histopathologic examination, whole lungs of mice were fixed and immersed in 4% paraformaldehyde neutral buffer overnight. Tissue sections of 4 μm thickness were cut and subjected to Haematoxylin and Eosin staining. Morphometric assessment was conducted with an Olympus BX53 inverted microscope (Olympus, NY, USA). Lung injury was scored blindly by a pathologist according to the degree of neutrophil infiltration, alveolar congestion, hemorrhage, and thickness of alveolar wall or hyaline membrane formation. The severity of lung injury was graded on a 0 to 4 scale: 0 = no injury; 1 = up to 25% injury of the field; 2 = up to 50% injury of the field; 3 = up to 75% injury of the field; and 4 = diffused injury [25].
2.3. Collection and analysis of bronchoalveolar lavage fluid (BALF)
BALF was collected by flushing the lungs with a tracheal cannula with 1 mL of ice-cold 1 × PBS for three times and centrifuged at 400 g for 5 min. Total cell number was calculated after removing the erythrocytes in the BALF by lysis buffer. 2 × 105 cells were smeared on a slide and stained with Giemsa regent (Nanjing Jiancheng Bio-engineering Institute, Nanjing, China) for cell differentiation, as previously described [26]. BALF protein concentrations were determined using the bicinchoninic acid (BCA) protein assay kit (Beyotime Biotechnology, Beijing, China). Lactate dehydrogenase (LDH) activity in BALF was determined with LDH Cytotoxicity Assay Kit (Nanjing Jiancheng Bioengineering Institute). Mouse cytokine enzyme-linked immunosorbent assay (ELISA) kits (Invitrogen, Carlsbad, CA, USA) were used to measure the levels of tumour necrosis factor (TNF)-α and interleukin (IL)-6 according to the manufacturer's instructions.
2.4. Lung permeability assay
The mice were injected intravenously with Evans blue (20 mg/kg, Sigma-Aldrich, USA) and sacrificed 1 h later, and the lung circulation was perfused by right ventricle puncture with 1 × PBS. Following drying at 56 °C for 48 h, lungs were weighed, and Evans blue dye was extracted in 1 mL of formamide (Sigma-Aldrich, USA) for 24 h at 60 °C. The supernatants were measured at 620 nm with a spectrophotometer (SpectraMax 5; Molecular Devices, Sunnyvale, CA, USA). Evans blue values were calculated as extracted Evans blue concentration comparing with a standard curve divided by the dry lung tissue weight.
2.5. Mitochondrial morphology detection
Mice were sacrificed at 24 h after treatments, and lungs were collected and fixed with 4% paraformaldehyde. Lung tissues were cut into 1 mm3 fragments and then processed with glutaraldehyde (2.5%) at 4 °C overnight. The tissues were processed through several chemical treatment steps (1% osmium tetroxide, distilled water and an ascending series of ethanol for dehydration). Then the samples were embedded in the mixture of propylene oxide and resin (1:1) overnight. After that, the samples were sliced into 100 nm slices and then stained using 4% uranyl acetate and 0.5% lead citrate. Finally, the ultrastructure of the mitochondria was observed under a transmission electron microscopy (FEI, Hillsboro, OR, USA).
2.6. Cell cultures and reagents
Immortalized HPMECs were purchased from Applied Biological Materials Incorporation (Richmond, BC, Canada). The cells were cultured in endothelial cell medium supplemented with 10% FBS, 100 U/mL of penicillin and 100 μg/mL of streptomycin at 37 °C with 5% CO2. Primary antibodies against vascular cell adhesion molecule-1 (VCAM-1), intercellular adhesion molecule-1 (ICAM-1), BAK, HO-1 , NF-κB P65, p-NF-κB P65 and mouse secondary antibody were purchased from Cell Signaling Technology (USA). Bcl2 antibody was obtained from Abcam (USA). β-Actin and Nrf2 antibodies were purchased from Huaan biotechology (China) and Arigo (Taiwan),respectively. GAPDH was obtained from Beyotime Institute of Biotechnology (China). Purified mouse anti-Cytochrome c and MitoTracker Red were bought from BD Pharmingen (USA) and Invitrogen (Carlsbad, CA, USA), respectively. Rabbit secondary antibody was bought from Beyotime Biotechnology. Pyrrolidinedithiocarbamate ammonium(PDTC)was purchase from Selleck (USA). MG132 and cycloheximide (CHX) were brought from Medchemexpress (USA).
2.7. Cell permeability assay
Cell monolayer permeability was measured by Transwell migration units (Corning, NY, USA), as previously described [27]. Briefly, 4–10 × 104 HPMECs were seeded per well and incubated for 48 h to be fully confluent before further treatments. 1 mg/mL FITC-labeled dextran (40 kDa; MilliporeSigma) was added to the upper chamber followed by a 30 min incubation before the fluorescence of 50 μL lower compartment sample was measured. Fluorescence was recorded at 520 nm, excited at 490 nm by spectrophotometer.
2.8. Immunoblot and immunofluorescence staining
Whole cell lysate was collected from immortalized HPMECs with 1 × RIPA lysis buffer containing 1 mmol/L phenylmethylsulfonyl fluoride and phosphatase inhibitor cocktail tablets (Roche, Basel, Switzerland). Concentrations of protein were determined by the BCA Protein Assay reagent (Beyotime Biotechnology). 50 μg protein of cell lysis was separated by SDS-PAGE electrophoresis and transferred to PVDF membrane (Millipore, Billerica, MA, USA). After blocking in Tris-buffered saline containing 0.1% Tween-20 with 5% fresh nonfat milk, the membranes were incubated with specific primary antibody (1: 1000) overnight at 4 °C followed by secondary antibody (1: 3000) incubation. The probed bands were visualized by chemiluminescence system (Syngene, Cambridge, UK).
For immunofluorescence staining, tissues or cells were fixed in 4% paraformaldehyde for 15 min before permeabilizing with 0.5% Triton X-100 for 20 min and blocking with 4% goat serum for 30 min. Samples were incubated with indicated Nrf2 or CD31 primary antibody (1: 100) at 4 °C overnightfollowed by mouse or rabbit secondary antibody (1: 2000) incubation. Images were collected under FV3000 fluorescent microscope (Olympus, USA).
2.9. Quantitative real-time PCR
Total cellular RNA extraction was performed using an ultrapure RNA kit (Cwbiotech, Beijing, China) according to the manufacturer's protocol. Synthesis of complementary DNA was performed with a cDNA synthesis system (Applied Biosystems, Foster City, CA, USA). cDNA amplification was performed using a QuantiFast SYBR Green PCR Kit (Qiagen, Hilden, Germany) in the CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). β-actin was used as an internal control. The comparative Ct method (2−ΔΔCt) was used to analyze data [28]. The sequences of primers for qPCR are as follows: β-actin forward:5′-GTATCCTGACCCTGAAGTACC-3’;
reverse: 5′-GAAGGTCTCAAACATGATCT- 3’;
HO-1 forward: 5′-TCTCTTGGCTGGCTTCCTTAC-3’;
reverse: 5′-GGCTTTTGGAGGTTTGAGACA-3’;
NQO-1 forward: 5′-TGCAGCGGCTTTGAAGAAGAAAG-3’;
reverse: 5′-TCGGCAGGATACTGAAAGTTCGC-3’.
2.10. Cell apoptosis assay
Cells were incubated with mitoQ (1 μM) for 2 h before they were treated with LPS (10 μg/mL) for 24 h. Detection of cell apoptosis was carried out according to the manufacturer's protocols using an FITC Annexin V Apoptosis Detection Kit (BD Biosciences, Franklin Lakes, NJ, USA). Briefly, the cells were washed twice with cold PBS before resuspending in binding buffer at a concentration of 1 × 106 cells/mL. Then cells were stained with 5 μL of FITC Annexin V and 5 μL of propidium iodide followed by incubation for 15 min at room temperature in the dark. Finally, apoptosis was analyzed with an ACEA NovoCyteTM (ACEA Biosciences, China).
2.11. Myeloperoxidase (MPO), malondialdehyde(MDA), superoxide dismutase (SOD), 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid (ABTS), ATP, TMRM and MitoSOX assays
Mitochondrial ATP activity was assayed using an ATP bioluminescence assay kit (Beyotime Institute of Biotechnology, China), according to the manufacturer's instructions. Mitochondrial transmembrane potential (△Ψm) in HPMECs was measured as the manufacturer's direction. Briefly, 25 nmol/L TMRM dye (Molecular Probes, Invitrogen, USA) was added to ECM medium for 30 min and assessed via spectrophotometer (SpectraMax 5; Molecular Devices, Sunnyvale, CA, USA). HPMECs were treated with salmon sperm DNA (10 μg/mL) for 2 h and then mitochondrial ROS was detected using MitoSOX (Invitrogen, USA) according to the manufacturer's instructions and measured via spectrophotometer. MPO, MDA, SOD and total antioxidant capacity were measured by MPO assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), MDA assay kits (Beyotime Institute of Biotechnology, China), SOD assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) and a rapid ABTS method (Beyotime Institute of Biotechnology, China), respectively.
2.12. Caspase 3 activity assay
A caspase 3 activity assay kit (Beyotime Institute of Biotechnology, China) was used according to the manufacturer's protocol. Briefly, after treatment with mitoQ (1 μM) and LPS (10 μg/mL), cell lysates were incubated with reaction buffer for 12 h at 37 °C, and the absorbance of the samples was measured at 405 nm with a spectrophotometer.
2.13. Molecular docking
The 3D structure of mitoQ was transformed from 2D version by Molecular Operating Environment (MOE v2015.1001, Chemical Computing Group Inc., Montreal, QC, Canada) through energy minimization. The X-ray structure of Nrf2 (PDB ID: 2LZ1) was obtained from Protein Data Bank (PDB). Before docking, the force field of AMBER10: EHT and the implied solvation model of Reaction Field (R-field) were selected. The protonation state of the protein and the orientation of the hydrogens were optimized using LigX, at a temperature of 300 K and pH 7.0 and energy minimized through MOE using default parameters. Molecular docking analyses were conducted using MOE software. The docking workflow was performed according to the “induced fit” protocol whereas side chains of the receptor pocket were allowed to interchange according to ligand conformations, with their limited positions. The weight used for tethering side chain atoms to their original positions was 10. All dock poses were scored using the ‘London dG’ scoring system and submitted to a refinement step based on molecular mechanics and rescored with the ‘GBVI/WSA dG’. GBVI/WSA dG, a forcefield-based scoring function, determines the binding free energy (kcal/mol) of the ligand from a given pose. The conformation with the lowest free energy of binding was selected as the most plausible binding mode. Molecular graphics were generated by PyMOL (www.pymol.org).
2.14. SiRNA transfection
Small interfering RNAs (siRNAs) were transfected into HPMECs with Hieff Trans™ Liposomal Transfection Reagent (Yeasen Biotechnology Co., Ltd., Shanghai, China) according to the manufacturer's instructions. SiRNAs were designed and synthesized by Shanghai GenePharma Co., Ltd (China). The siRNA sequences targeting human Nrf2 were as follows: sense: 5′-GACAGAAGUUGACAAUUAUTT-3′;
and antisense, 5′-AUAAUUGUCAACUUCUGUCTT-3’.
The siRNA sequences targeting human Maff were sense: 5′-CCAGCAAAGCUCUAAAGAUTT-3′;
and antisense: 5′-CCAGCAAAGCUCUAAAGAUTT-3’.
The scramble control sequences were sense: 5′-UUCUCCGAACGUGUCACGUTT-3′;
and antisense: 5′-ACGUGACACGUUCGGAGAATT-3’.
2.15. Statistical analysis
All quantitative data were shown as means ± SEM and performed with Prism (GraphPad, La Jolla, CA, USA). Differences between samples were analyzed using Student's t-test. Survival curve analysis was performed by the log rank test. Values of P < 0.05 were considered as statistically significant.
3. Results
3.1. MitoQ alleviated the oxidant damage, inflammation and lung tissue injury in mice with ALI
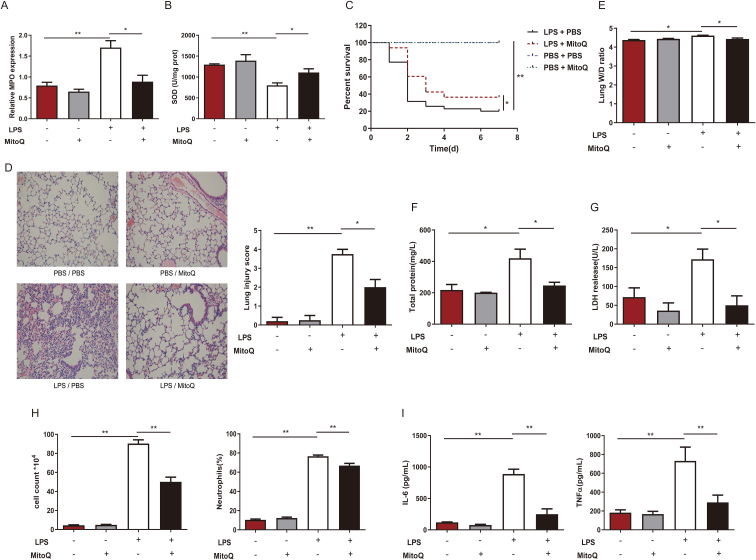
To investigate the oxidative status during acute inflammation situation, we detected two specific indicators, MPO and SOD, in an LPS-induced ALI mouse model. We found remarkable upregulation of MPO and decrease in SOD, in lungs of mice with ALI, which indicated oxidative stress. And these were partially reversed by mitoQ pretreatment, which suggested mitoQ might exert oxidant balance preservation effect during an inflammatory response (Fig. 1A and B). To further study the role antioxidants played in inflammatory situation, we pretreated mice with mitoQ or vehicle and injected intratracheally with a lethal dose of LPS for survival rate calculation or a sublethal dose for injury evaluation. We found ALI mice administrated with mitoQ shown much higher survival rate compared to LPS stimulated group (Fig. 1C). Furthermore, mitoQ administration significantly reduced the inflammation in ALI mice. Pulmonary histopathological examinations demonstrated that low dose of LPS induced remarkable morphological injury, including interstitial edema, congestion and infiltration of inflammatory cells into the parenchyma and alveolar spaces of lung, which was significantly reduced in mitoQ group. Lung injury scores elevated by LPS were also decreased by mitoQ (Fig. 1D). Indicators of alveolar endothelial barrier and cellular damage, lung wet-to-dry ratio, concentrations of total protein and LDH in BALF increased remarkably after LPS treatment, but were partially reversed by mitoQ pretreatment (Fig. 1E–G). Notable decreases in cell count, proportion of neutrophil cells as well as inflammatory factors IL-6 and TNF-α in BALF were also observed in mice after mitoQ treatment (Fig. 1H and I). These results suggested that mitoQ was capable of alleviating the oxidative stress and lung inflammation during ALI.
Fig. 1.
MitoQ alleviated the oxidant damage, inflammation and lung tissue injury in mice with ALI. (A–I) C57/BL6 mice were i. t. with a lethal dose (50 mg/kg, n = 10–35 mice/group) (C) or a sublethal dose (5 mg/kg, n = 3–7 mice/group) (A-B, D-I) of LPS after i. p. with or without mitoQ(5 mg/kg) 30 min earlier. (A–B) MPO and SOD were determined in lung tissue. (C) Mice survival rate was observed every 24 h up to 7 d. (D) Micrographs of haematoxylin and eosin staining and lung injury score in lung sections at 12 h following stimulation were observed and analyzed (Magnification, × 200). (E) Wet/dry ratio of lung tissue was measured. (F–I) Total protein, LDH, cell counts and proportion of neutrophils, and levels of IL-6, TNF-α were determined in the BALF. Quantitative data were shown as mean ± SEM, *P < 0.05; **P < 0.01.
3.2. MitoQ regulated the function of endothelium and inhibited hyperpermeability of endothelium in mice with ALI
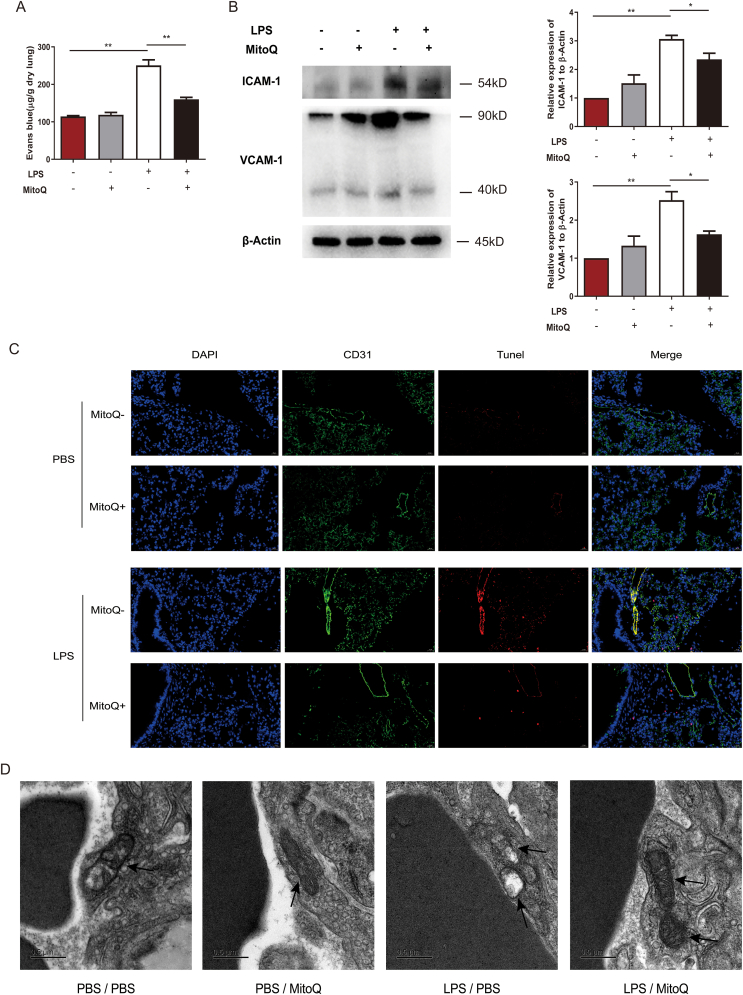
Since dysfunction of microvascular barriers is considered to be an important pathophysiology procedure in ALI, we then determined the permeability of lung after LPS treatment with mitoQ or vehicle and found mitoQ prevented Evans Blue leaking from microvascular of the lungs (Fig. 2A). To assess the function of endothelial cells, we detected the expression of some adhesion molecules such as vascular cell adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1) and found an upregulation of these protein expression in ALI mice, which was obviously inhibited by mitoQ treatment (Fig. 2B). To explore the reason of dysfunction of endothelial barrier, the apoptosis of endothelium was detected. The results of immunofluorescence revealed that mitoQ significantly decreased the proportion of apoptosis of CD31+ endothelial cells in ALI mice (Fig. 2C). In endothelial cells of lungs from control group, mitochondria were long spindles with dense matrix and well-developed cristae. In contrast, mitochondria in LPS treated group represented in swelling forms, lower dense matrix and missing cristae, which could be partially reversed by pretreatment of mitoQ (Fig. 2D). In general, mitoQ might regulate the function and inhibit the apoptosis of endothelial cells through maintaining the quality of mitochondria of endothelial cells.
Fig. 2.
MitoQ inhibited hyperpermeability, regulated the function and reduced apoptosis of endothelial cells in mice with ALI. C57/BL6 mice were i. t. with a dose (5 mg/kg) of LPS after i. p. with or without mitoQ (5 mg/kg) 30 min earlier, n = 3–4 mice/group. (A) Evans blue concentration was determined in lung tissue. (B) The expression of VCAM-1, ICAM-1 and β-Actin were analyzed by immunoblot. (C) TUNEL, CD31 and DAPI were detected by immunofluorescence (Magnification, × 400). (D) Mitochondrial ultrastructure (black arrow) was observed under a transmission electric microscopy (Magnification, × 17,000). Values were shown as mean ± SEM, *P < 0.05; **P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. MitoQ eliminated excessive ROS and protected the function of mitochondria in LPS-stimulated HPMECs
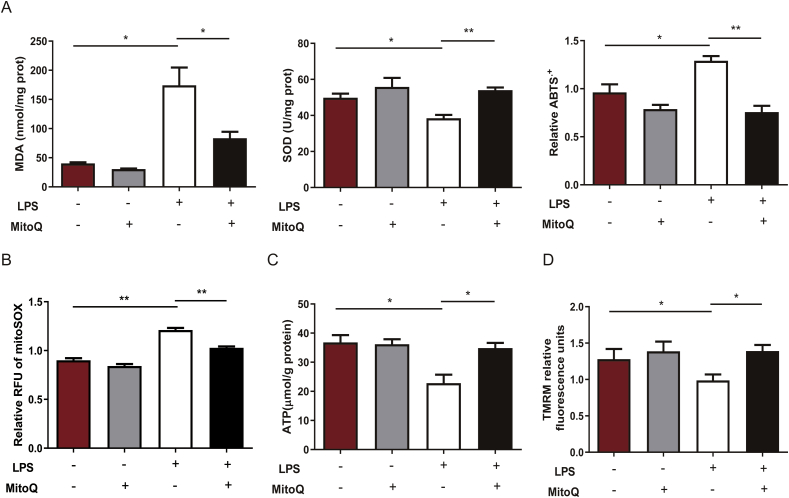
To investigate whether mitoQ has consistent protective effect on endothelial cells in in vitro model, we incubated HPMECs with LPS (10 μg/mL) for 24 h after administrated with mitoQ (1 μM) or PBS, and measured specific indicators of ROS production and function of mitochondria. MDA and values of ABTS assay were both upregulated while SOD was downregulated in LPS-stimulated HPMECs, which suggested an increase in ROS production. However, this effect was notably abolished by mitoQ (Fig. 3A). In addition, ROS produced in mitochondria, reflected by relative RFU of mitoSOX, was inhibited by mitoQ in LPS stimulated HPMECs (Fig. 3B). ATP production in HPMECs was significantly reduced by LPS accompanied with the decrease of mitochondrial membrane potential detected by TMRM, which were both restored by mitoQ (Fig. 3C and D). These results demonstrated that mitoQ exerted protective effects on mitochondrial homeostasis in inflammatory situation.
Fig. 3.
MitoQ inhibited oxidative stress and protected the function of mitochondria in LPS-stimulated HPMECs. HPMECs were stimulated with LPS (10 μg/mL) for 24 h after incubation with 1 μM MitoQ for 2 h. (A) MDA, SOD and ABTS were determined. (B–D) MitoSOX, ATP and TMRM were measured, respectively. The data were presented as mean ± SEM, *P < 0.05; **P < 0.01.
3.4. MitoQ reduced the mitochondrial pathway apoptosis of HPMECs induced by LPS
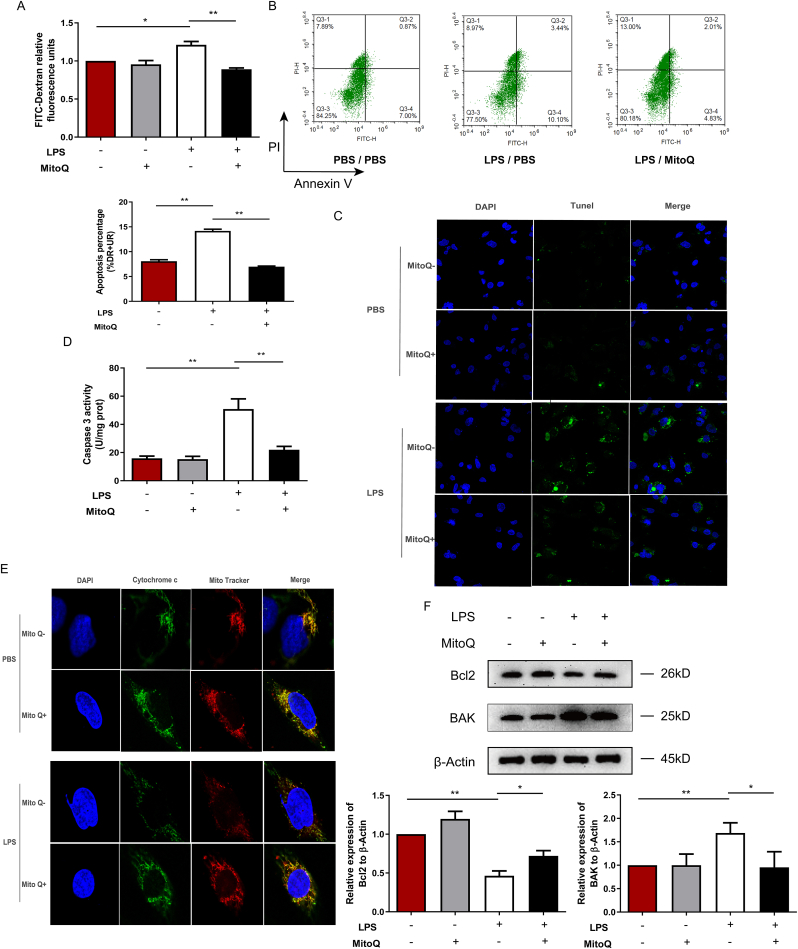
To further investigate the role of mitoQ in vitro, we stimulated HPMECs with LPS after treatment with mitoQ or PBS, and then detected the cell monolayer permeability and apoptosis. These results revealed that mitoQ remarkably protected HPMECs from hyperpermeability and apoptosis induced by LPS (Fig. 4A–C). To explore the mechanism underlying, we determined the activity of caspase 3, which was upregulated by LPS stimulation, but also reduced by mitoQ (Fig. 4D). Furthermore, we detected the colocalization of mitochondria and cytochrome c. LPS induced cytochrome c releasing from mitochondria, which was significantly reversed by mitoQ (Fig. 4E). Consistently, the expression of anti-apoptotic factor Bcl2 increased while pro-apoptotic factor BAK decreased, which will reduce permeabilization of the outer mitochondria membrane, in mitoQ/LPS group, compared with LPS treated cells (Fig. 4F). Hyperpermeability of the outer mitochondria membrane will further induce the release of cytochrome c from mitochondria, and activate the caspases cascade which will promote the cell apoptosis [29,30]. Our results showed mitoQ obviously reversed this procedure.
Fig. 4.
MitoQ prevented hyperpermeability and apoptosis of HPMECs. HPMECs were incubated with 1 μM mitoQ for 2 h before stimulation with LPS (10 μg/mL) for 24 h. (A) Relative fluorescence units were detected in lower chambers after incubation with FITC-Dextran for 1 h. (B–C) The apoptosis of HPMECs was measured by flow cytometry and Tunel staining (Magnification, × 400). (D) The activity of Caspase 3 was measured. (E) Cytochrome c and Mito Tracker were observed under a fluorescent microscope (Magnification, × 1200). (F) Bcl2 and BAK were measured by Western Blot. Quantitative data are shown as mean ± SEM, *P < 0.05; **P < 0.01.
3.5. MitoQ attenuated degradation of Nrf2 and promoted nuclear translocation of Nrf2
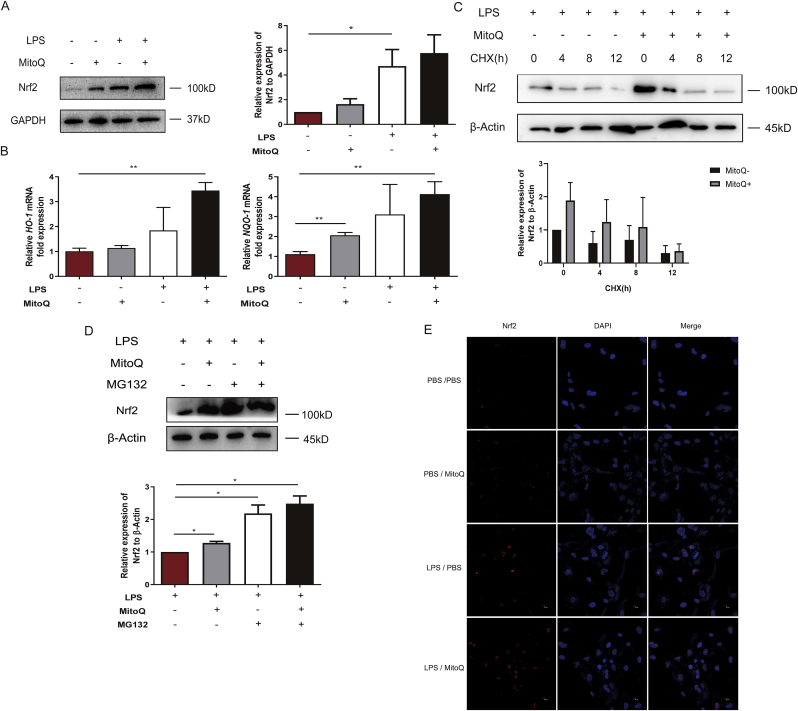
In order to explore the mechanism of the protective function of mitoQ in LPS induced HPMECs, we proceeded RNA-sequencing of HPMECs treated with LPS after PBS or mitoQ administration and identified 20 genes whose expression changes most significantly (Fig. S1). By consulting the literature, we focused on MafF, a small protein, which could interact with Nrf2. The expression of Nrf2, a transcription factor inducing the expression of AREs, did not changed significantly in mRNA level (Fig. S2). However, as shown in Fig. 5A, B and 6A compared to HPMECs simply exposed to LPS, pretreatment with mitoQ promoted the expression of Nrf2 in protein level and downstream AREs (HO-1, NQO-1). Then we sought to explore the mechanism of Nrf2 upregulation. HPMECs were treated with cycloheximide (CHX) after pretreated with LPS and mitoQ, but the influence of mitoQ on Nrf2 wasn't eliminated (Fig. 5C). In addition, proteasome inhibitor MG132 could significantly upregulate the expression of Nrf2 with or without mitoQ (Fig. 5D). These results suggested mitoQ mediated the proteasomal degradation of Nrf2. Moreover, immunofluorescence staining showed that mitoQ remarkably promoted the nucleus translocation of Nrf2 (Fig. 5E).
Fig. 5.
MitoQ attenuated degradation of Nrf2 and promoted its nuclear translocation. HPMECs were stimulated with LPS (10 μg/mL) for 24 h after incubation with 1 μM mitoQ for 2 h. (A) The expression of Nrf2 was determined using immunoblot. (B) The expression of HO-1 and NQO-1 were determined by QPCR. (C–D) HPMECs were stimulated with LPS (10 μg/mL) for 24 h following incubation with 1 μM mitoQ for 2 h, then treated with CHX (1 μM) for indicated times respectively or MG132 (1 μM). The expression of Nrf2 was determined using immunoblot. (E) After HPMECs were treated with mitoQ and LPS as mentioned above, Nrf2 translocation was detected by immunofluorescence. The data were presented as mean ± SEM, *P < 0.05; **P < 0.01.
3.6. MitoQ and PDTC collabotared to up-regulate the expression of Nrf2
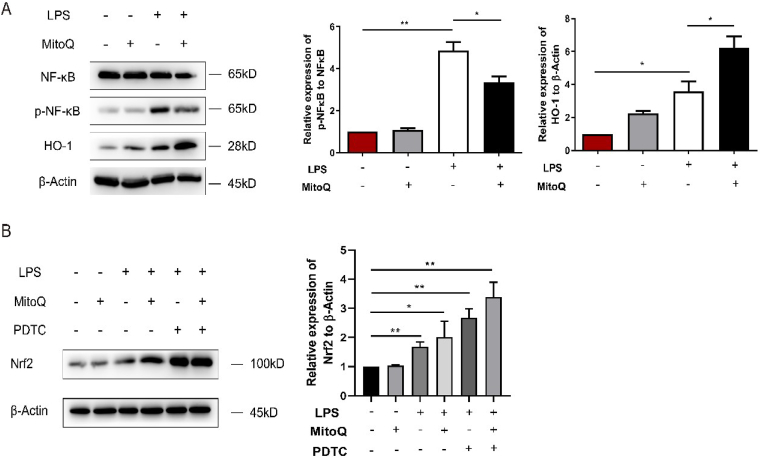
To further investigate the influence of mitoQ on the inflammatory process, we detected the phosphorylation of NF-κB P65 and found that mitoQ was capable of downregulating the activation of NF-κB P65. The protein level of HO-1 was upregulated by mitoQ (Fig. 6A). Furthermore, inhibiting the activation of the NF-κB by PDTC increased the expression of Nrf2, and this function of PDTC could be collaborated with mitoQ (Fig. 6B).
Fig. 6.
MitoQ and PDTC collabotared to up-regulate the expression of Nrf2. (A) HPMECs were incubated with LPS (10 μg/mL) for 24 h after treatment with 1 μM mitoQ for 2 h, and the expression of NF-κB, HO-1 and p-NF-κB were detected by immunoblot. (B) HPMECs were pretreated with PDTC (50 mM) for 1 h, and then incubated with 1 μM mitoQ for 2 h before stimulation with LPS (10 μg/mL). The expression of Nrf2 was detected using immunoblot. The data were shown as mean ± SEM, *P < 0.05; **P < 0.01.
3.7. MitoQ inhibited apoptosis of HPMECs induced by LPS exposure partially via Nrf2 and MafF pathway
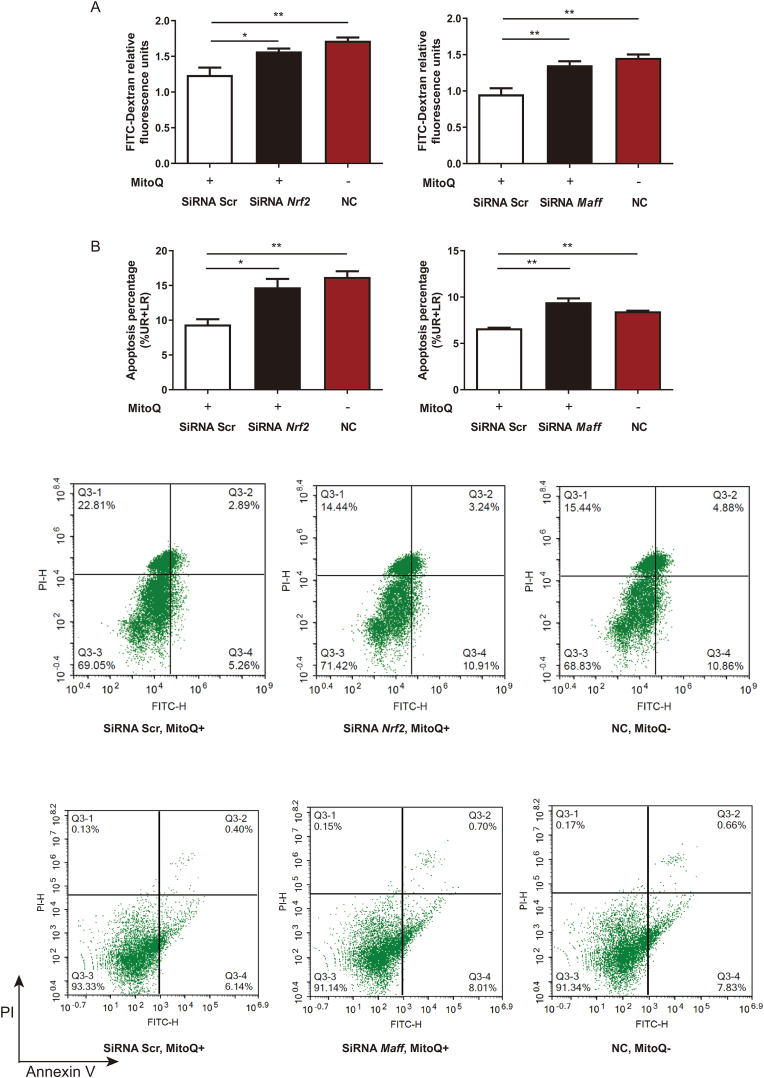
To explore whether mitoQ exerts its protective function through MafF and Nrf2, we knocked down the expression of Maff and Nrf2 by siRNA (Fig. S3). Our current data suggested that the effect of mitoQ downregulating the permeability of LPS-treated cells was abolished by either Maff or Nrf2 siRNA transfection (Fig. 7A). Similarly, the apoptosis proportion of cells increased by silence of Maff or Nrf2 (Fig. 7B). These results demonstrated that mitoQ exerted its inhibition of hyperpermeability and apoptosis in HPMCEs partially via MafF-Nrf2 pathway.
Fig. 7.
MitoQ inhibited apoptosis in HPMECs subjected to LPS exposure partially via Nrf2 and MafF. HPMECs were transfected with Nrf2, Maff or scramble siRNA 24 h before treated with mitoQ (1 μM) for 2 h and LPS (10 μg/mL) for 24 h. (A) Relative fluorescence units were detected after incubation with FITC-Dextran for 1 h. (B) Apoptosis of HPMECs was measured by flow cytometry. NC: negative control. Quantitative data were shown as mean ± SEM, *P < 0.05; **P < 0.01.
3.8. MitoQ formed a suitable steric complementarity with the binding site of Nrf2
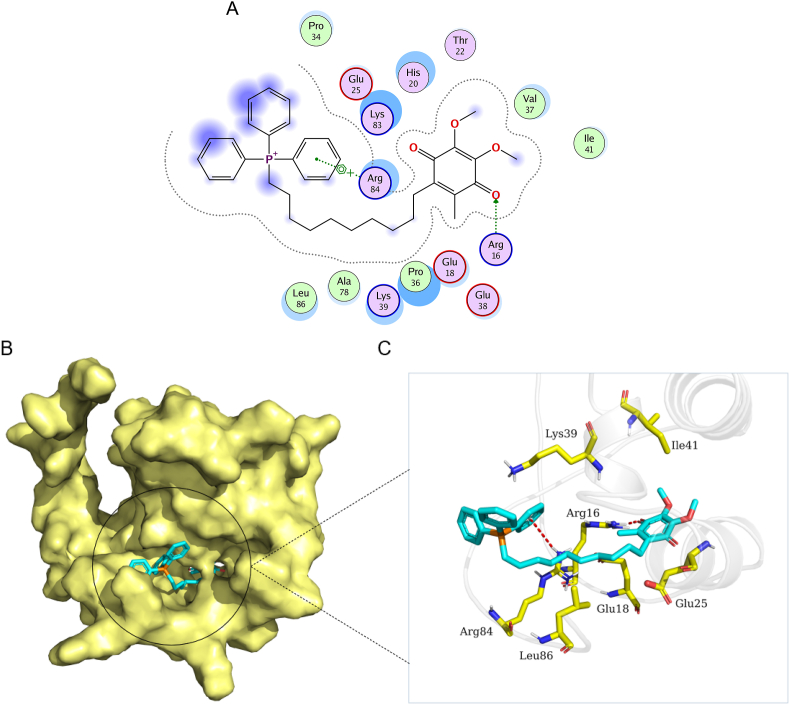
To further investigate the interaction between mitoQ and Nrf2, we did a molecular docking. The docking score of mitoQ with Nrf2 was -6.81 (kcal/mol). The binding mode of mitoQ with protein Nrf2 was illustrated in Fig. 8. MitoQ has made a steric complementarity bridge with the binding site of Nrf2 where hydrogen bond and sigma-pi conjugation interactions were formed. One oxygen atom of mitoQ was observed as hydrogen bond acceptor and formed a hydrogen bond with the side chain nitrogen atom (Arg16 residue). It can be seen that this binding site (Arg16) of nitrogen atom forms sigma-pi conjugation interaction with one benzene ring of mitoQ. Van der Waals (VDW) interactions were also formed among mitoQ and the surrounding residues (Fig. 8). Therefore, these interactions mainly contribute to a promising potential binding energy between mitoQ with protein Nrf2.
Fig. 8.
MitoQ was capable of forming conformation with Nrf2. (A) The 2D binding mode of mitoQ and Nrf2. (B) The binding model of mitoQ on molecular surface of Nrf2. MitoQ was colored in cyan and the molecular surface of Nrf2 was colored in pale yellow. (C) The 3D binding mode of mitoQ and Nrf2. MitoQ was colored in cyan and the surrounding residues of Nrf2 in the binding pockets were colored in yellow, the backbone of the receptor was depicted as white cartoon with transparency. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.9. Nrf2 deficiency abolished the protective function of mitoQ in mice with ALI
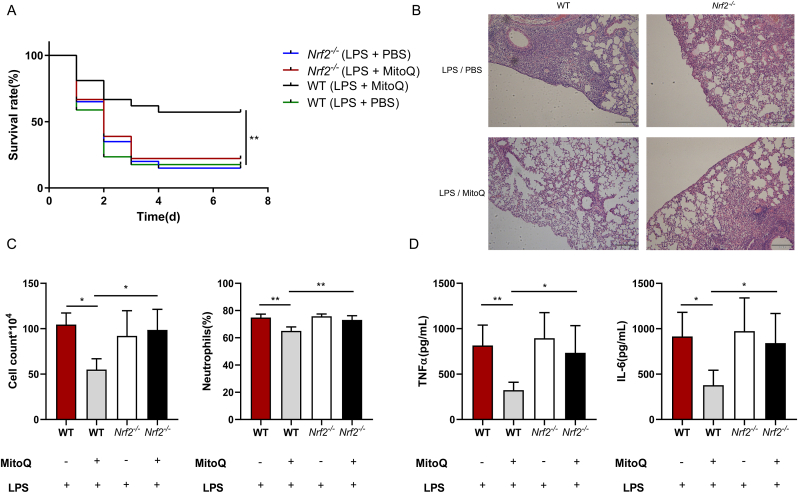
In order to confirm the role of Nrf2 in the mice model of ALI, both WT and Nrf2 deficient mice were injected intratracheally with LPS after mitoQ or PBS administration. The survival rate of Nrf2 deficient mice represented no significant difference between LPS/mitoQ and LPS/PBS treated group, both similar to WT LPS/PBS group (Fig. 9A). Analogously, HE staining of Nrf2 deficient mice in both groups upon LPS stimulation showed similar pulmonary morphology changes, including alveolar infiltration of inflammatory cells, alveolar wall thickening and interstitial edema (Fig. 9B). When treated with LPS, Nrf2 deficient mice pretreated with mitoQ showed increased cell count and neutrophil proportion compared with WT mitoQ group, accompanied with elevated inflammation factors, including IL-6 and TNF-α, but similar to WT PBS group and Nrf2 PBS group (Fig. 9C and D). These results suggested that mitoQ protected ALI mice from extensive inflammatory response through Nrf2 pathway.
Fig. 9.
The protective function of mitoQ from lung tissue injury in mice with ALI was abolished by Nrf2 deficiency. (A) C57/BL6 mice were i. t. with a lethal dose (50 mg/kg) of LPS after i. p. with or without mitoQ (5 mg/kg) 30 min earlier. Mice survival was observed every 24 h up to 7 d, n = 17–21 mice/group. (B–D) C57/BL6 mice were i. t. with a sublethal dose (5 mg/kg) of LPS for 12 h after i. p. with or without mitoQ (5 mg/kg) 30 min earlier, n = 4–5 mice/group. (B) Lungs sections stained with haematoxylin and eosin were observed using a microscope (Magnification, × 200). (C) Cell counts and proportion of neutrophils were recorded. (D) The levels of IL-6 and TNF-α were determined in the BALF. Values were shown as mean ± SEM, *P < 0.05; **P < 0.01.
4. Discussion
Our current study demonstrated that LPS activated excessive amounts of oxidative agents and these promoted the oxidative stress in ALI mouse model, which could be mitigated by the mitochondrial target antioxidant agent known as mitoQ. Additionally, we showed that administration of mitoQ repressed pulmonary inflammation and barrier dysfunction in ALI via anti-oxidative stress. Furthermore, the possible underlying mechanisms of mitoQ might be related to ROS reduction and the mitochondrial protection in endothelial cells via Nrf2-MafF/ARE signaling pathway, which inhibited the endothelial cell apoptosis in extensive inflammatory circumstances. Beside of this, molecular docking implicated that mitoQ was capable of forming stable conformation with Nrf2, which justified a role for mitoQ in reducing oxidative injury in ALI/ARDS via Nrf2-MafF/ARE axis.
It has been noted that hyperpermeability of microvascular barriers, accounting for extravascular accumulation of leukocytes and protein-rich edema fluid, is the pivotal pathophysiology procedure in ALI/ARDS [31]. Impairment of alveolar epithelial and endothelial permeability involved a variety of important molecular mechanisms, including the activation of myosin light chain kinase (MLCK), RhoA and tyrosine kinases; increase of calcium influx; and apoptosis of the endothelial cells, etc. [32,33]. In most eukaryotic cells, mitochondria play a key role in cellular energy production and programmed cell death [34,35]. Dysfunction of mitochondria involves overproduction of ROS, insufficient generation of ATP and increases in mitochondrial fragmentation. These alternative markers later induce apoptosis in cells. Previously, it was reported that bone marrow–derived stromal cells (BMSCs) protected against ALI by restituting alveolar bioenergetics through Cx43-dependent alveolar attachment and mitochondrial transfer [36]. Controlling of mitochondrial homeostasis has been recognized as a potential treatment strategy of ALI/ARDS, but the clear mechanism of action is not yet to reveal. Thus, more studies should be performed to find potential target molecules for drug development.
Numerous studies have reported that a few antioxidant agents like vitamin E and coenzyme Q10 have disappointing clinical effect such as poor uptake into the body as well as limited delivery to the mitochondria [12,37]. To overcome this limitation, mitoQ, a mitochondria targeted antioxidant [38], was implicated in this study. In our in vivo study, LPS triggered mitochondrial oxidative stress, which resulted in severe mitochondrial injury in lung. And this was markedly repressed by mitoQ. Treatment of mitoQ alleviated inflammation in ALI mice, which was reflected in decreased amounts of inflammatory cells and inflammatory mediators as TNF-α and IL-6 in BALF (Fig. 1). Moreover, mitoQ showed its protective activity on hyperpermeability of microvascular barriers and improved the quality of mitochondria (Fig. 2). Consistent with the in vivo study, LPS stimulated ROS production in endothelial cells and disrupted the function of mitochondria, whereas administration of mitoQ dramatically inhibited the generation of ROS and improved the condition of mitochondria (Fig. 3). In the meantime, mitoQ largely reduced the hyperpermeability of HPMECs monolayer and inhibited mitochondrial pathway of apoptotic cell death via regulating the expression of anti-apoptosis protein Bcl-2 and pro-apoptosis BAK, and reduced the release of cytochrome c from mitochondria and the activity of caspase 3 in endothelial cells (Fig. 4). These results are consistent with those of previous reports where Sha Chen and their research group regarding inhibition of ROS and excess autophagy in CSE-induced HUVEC injury by NFκB/NLRP3 pathway [39]. In another intestinal ischemia reperfusion (I/R) model study, pretreatment of mitoQ downregulated I/R-induced oxidative stress, inhibited epithelial apoptosis, and stabilized the intestinal barrier compared to controls [40]. Thus, mitoQ exerts protective effect on I/R-induced intestinal barrier dysfunction [40]. Recently Ruirui Li and his colleague revealed that mitoQ protected lung function and inhibited cell apoptosis in lung tissue by activating the PI3K/Akt/GSK-3 beta/mTOR pathway [41]. Our findings indicate that mitoQ plays an efficient role in LPS induced acute lung injury via inhibiting mitochondrial ROS (mtROS) production in both in vivo and in vitro.
Activation of Nrf2/ARE signaling is a crucial process in protecting cells from oxidative insults [42]. However, there remain controversies in the mechanism how Nrf2 is exactly activated. Several studies reported that the gene expression of Nrf2 might be determined by complex I activity via an ERK5-myocyte enhancer factor 2 (MEF2) signaling pathway independent of ROS [43,44]. While other studies reported changes in mtROS generation actually altered the cellular response, where changes of mtROS actually influenced the activation of Nrf2 through unknown mediators or signaling cascades [45,46]. In a recent study from Ping Wang et al. the stability of Nrf2 was modulated by kinases Mst1 and Mst2 (Mst1/2) which can sense site-specific ROS release and maintain cellular redox homeostasis. Activated Mst1/Mst2 by mtROS could phosphorylate Keap1 at four Ser/Thr residues in the N-terminus, which inactivated Keap1 and decreased the degradation of Nrf2. Thus, these results suggested that mtROS activated Nrf2 mainly by indirect and kinase-dependent way [47]. Accumulating evidence also suggests that the Nrf2/ARE pathway reduces apoptosis and alleviates the inflammatory response, resulting in a significant improvement in tissue retention and beneficial role in coordination [48]. It is also reported that oxidative signaling and Nrf2 were tightly linked with cellular responses [49]. Previous studies have revealed mitoQ exerted its protective effects on diseases like traumatic brain injury [24], intestinal ischemia reperfusion [40] and diabetic kidney disease [17] through regulation of Nrf2. In db/db mice, mitoQ partially rectified mitochondrial dysfunctions, such as defective mitophagy, mtROS overexpression and mitochondrial fragmentation in tubular cells and inhibited tubular cells apoptosis [17]. In in vitro study mitoQ decreased deficiency of mitophagy, mitochondrial dysfunction and apoptosis in high glucose induced HK-2 cells via Nrf2/PINK pathway [17]. In this study, we found that the protective functions of mitoQ in ALI were eliminated in Nrf2 deficient mice, compared to WT group. Consistently, when MafF and Nrf2 were knocked down by siRNA in HPMECs, mitoQ was not able to attenuate the hyperpermeability of monolayer as well as cell apoptosis in HPMECs. Therefore, we speculated that though eliminating superoxide after alteration by complex II was one of the important effects of mitoQ, and stabilizing Nrf2 might be another dominant antioxidative function of mitoQ. In general, our results showed a potent evidence for the treatment of ALI by mitoQ through Nrf2-MafF/ARE signaling pathway.
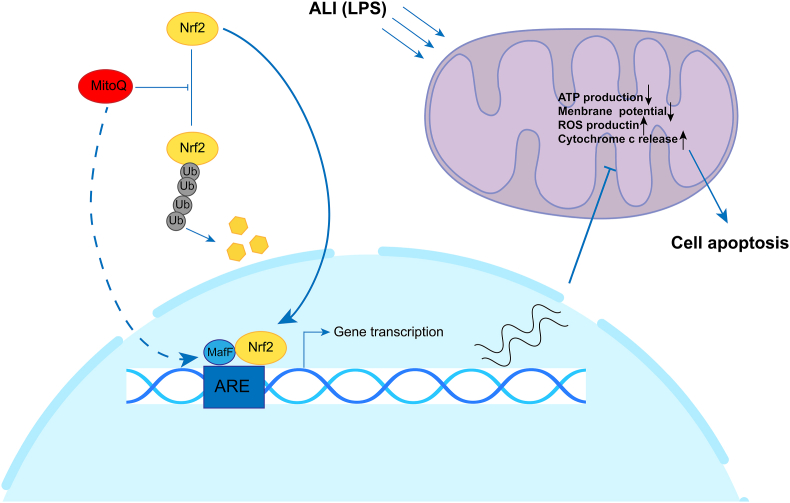
In summary, we explored mitoQ and its attenuation of LPS induced inflammation in in vitro and in vivo model. The mechanisms underlying these effects may involve reducing mitochondrial ROS, maintaining the mitochondrial homeostasis, inhibiting apoptosis via Nrf2-MafF/ARE signaling in endothelial cells (Fig. 10). These findings identify a novel role of mitoQ as a potential substance for treatment of ALI/ARDS.
Fig. 10.
Possible molecular mechanism of mitoQ protecting the endothelial cell from injury under the inflammatory condition. In simulation by LPS, mitoQ up-regulates the MafF expression and inhibits the proteasome pathway degradation of Nrf2. Simultaneously, mitoQ significantly promotes the nucleus translocation of Nrf2, which enhances the ARE transcription. The downstream products of ARE greatly reduce the over-production of ROS stimulated by the inflammatory circumstance, resulting in the restoration of mitochondrial homeostasis and inhibition of mitochondrial pathway cell apoptosis.
Funding source
The project was supported by National Natural Science Foundation of China (No. 81873465).
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in the manuscript entitled, “MitoQ protects against hyperpermeability of endothelium barrier in acute lung injury via a Nrf2-dependent mechanism”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101936.
Contributor Information
Jingyan Xia, Email: xiajingyan2018@zju.edu.cn.
Feng Xu, Email: xufeng99@zju.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Shah R.D., Wunderink R.G. Viral pneumonia and acute respiratory distress syndrome. Clin. Chest Med. 2017;38:113–125. doi: 10.1016/j.ccm.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthay M.A., Zemans R.L., Zimmerman G.A., Arabi Y.M., Beitler J.R., Mercat A., Herridge M., Randolph A.G., Calfee C.S. Acute respiratory distress syndrome. Nat. Rev. Dis. Prim. 2019;5 doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthay M.A., McAuley D.F., Ware L.B. Clinical trials in acute respiratory distress syndrome: challenges and opportunities. Lancet Respir. Med. 2017;5:524–534. doi: 10.1016/S2213-2600(17)30188-1. [DOI] [PubMed] [Google Scholar]
- 4.Szabari M.V., Takahashi K., Feng Y., Locascio J.J., Chao W., Carter E.A., Vidal Melo M.F., Musch G. Relation between respiratory mechanics, inflammation, and survival in experimental mechanical ventilation. Am. J. Respir. Cell Mol. Biol. 2019;60:179–188. doi: 10.1165/rcmb.2018-0100OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiśniewski M., Bieniek A., Roszek K., Czarnecka J., Bolibok P., Ferrer P., da Silva I., Terzyk A.P. Cystine-based MBioF for maintaining the antioxidant–oxidant balance in airway diseases. ACS Med. Chem. Lett. 2018;9:1280–1284. doi: 10.1021/acsmedchemlett.8b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cui L., Zhou Q., Zheng X., Sun B., Zhao S. Mitoquinone attenuates vascular calcification by suppressing oxidative stress and reducing apoptosis of vascular smooth muscle cells via the Keap1/Nrf2 pathway. Free Radic. Biol. Med. 2020;161:23–31. doi: 10.1016/j.freeradbiomed.2020.09.028. [DOI] [PubMed] [Google Scholar]
- 7.Choi J.S., Lee H.S., Seo K.H., Na J.O., Kim Y.H., Uh S.T., Park C.S., Oh M.H., Lee S.H., Kim Y.T. The effect of post-treatment N -acetylcysteine in LPS-induced acute lung injury of rats. Tuberc. Respir. Dis. 2012;73:22–31. doi: 10.4046/trd.2012.73.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arkovitz M.S., Wispé J.R., Garcia V.F., Szabó C. Selective inhibition of the inducible isoform of nitric oxide synthase prevents pulmonary transvascular flux during acute endotoxemia. J. Pediatr. Surg. 1996;31:1009–1015. doi: 10.1016/s0022-3468(96)90075-5. [DOI] [PubMed] [Google Scholar]
- 9.Sarma J.V., Ward P.A. Oxidants and redox signaling in acute lung injury. Comp. Physiol. 2011;1:1365–1381. doi: 10.1002/cphy.c100068. [DOI] [PubMed] [Google Scholar]
- 10.Grimsrud P.A., Xie H., Griffin T.J., Bernlohr D.A. Oxidative stress and covalent modification of protein with bioactive aldehydes. J. Biol. Chem. 2008;283:21837–21841. doi: 10.1074/jbc.R700019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erdinçler D.S., Seven A., Inci F., Beğer T., Candan G. Lipid peroxidation and antioxidant status in experimental animals: effects of aging and hypercholesterolemic diet. Clin. Chim. Acta. 1997;265:77–84. doi: 10.1016/s0009-8981(97)00106-x. [DOI] [PubMed] [Google Scholar]
- 12.Murphy M.P., Smith R.A.J. Drug delivery to mitochondria: the key to mitochondrial medicine. Adv. Drug Deliv. Rev. 2000;41:235–250. doi: 10.1016/S0169-409X(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 13.Kelso G.F., Porteous C.M., Coulter C.V., Hughes G., Porteous W.K., Ledgerwood E.C., Smith R.A.J., Murphy M.P. Selective targeting of a redox-active ubiquinone to mitochondria within cells: antioxidant and antiapoptotic properties. J. Biol. Chem. 2001;276:4588–4596. doi: 10.1074/jbc.M009093200. [DOI] [PubMed] [Google Scholar]
- 14.Maroz A., Anderson R.F., Smith R.A.J., Murphy M.P. Reactivity of ubiquinone and ubiquinol with superoxide and the hydroperoxyl radical: implications for in vivo antioxidant activity. Free Radic. Biol. Med. 2009;46:105–109. doi: 10.1016/j.freeradbiomed.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 15.James A.M., Sharpley M.S., Manas A.-R.B., Frerman F.E., Hirst J., Smith R.A.J., Murphy M.P. Interaction of the mitochondria-targeted antioxidant MitoQ with phospholipid bilayers and ubiquinone oxidoreductases. J. Biol. Chem. 2007;282:14708–14718. doi: 10.1074/jbc.M611463200. [DOI] [PubMed] [Google Scholar]
- 16.Snow B.J., Rolfe F.L., Lockhart M.M., Frampton C.M., O’Sullivan J.D., Fung V., Smith R.A.J., Murphy M.P., Taylor K.M. A double-blind, placebo-controlled study to assess the mitochondria-targeted antioxidant MitoQ as a disease-modifying therapy in Parkinson’s disease. Mov. Disord. 2010;25:1670–1674. doi: 10.1002/mds.23148. [DOI] [PubMed] [Google Scholar]
- 17.Xiao L., Xu X., Zhang F., Wang M., Xu Y., Tang D., Wang J., Qin Y., Liu Y., Tang C., He L., Greka A., Zhou Z., Liu F., Dong Z., Sun L. The mitochondria-targeted antioxidant MitoQ ameliorated tubular injury mediated by mitophagy in diabetic kidney disease via Nrf2/PINK1. Redox Biol. 2017;11:297–311. doi: 10.1016/j.redox.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escribano-Lopez I., Diaz-Morales N., Rovira-Llopis S., de Marañon A.M., Orden S., Alvarez A., Bañuls C., Rocha M., Murphy M.P., Hernandez-Mijares A., Victor V.M. The mitochondria-targeted antioxidant MitoQ modulates oxidative stress, inflammation and leukocyte-endothelium interactions in leukocytes isolated from type 2 diabetic patients. Redox Biol. 2016;10:200–205. doi: 10.1016/j.redox.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adlam V.J., Harrison J.C., Porteous C.M., James A.M., Smith R.A.J., Murphy M.P., Sammut I.A. Targeting an antioxidant to mitochondria decreases cardiac ischemia‐reperfusion injury. FASEB. J. 2005;19:1088–1095. doi: 10.1096/fj.05-3718com. [DOI] [PubMed] [Google Scholar]
- 20.Lowes D.A., Thottakam B.M.V., Webster N.R., Murphy M.P., Galley H.F. The mitochondria-targeted antioxidant MitoQ protects against organ damage in a lipopolysaccharide–peptidoglycan model of sepsis. Free Radic. Biol. Med. 2008;45:1559–1565. doi: 10.1016/j.freeradbiomed.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc. Natl. Acad. Sci. Unit. States Am. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho H.Y., Reddy S.P., Kleeberger S.R. Nrf2 defends the lung from oxidative stress. Antioxidants Redox Signal. 2006;8:76–87. doi: 10.1089/ars.2006.8.76. [DOI] [PubMed] [Google Scholar]
- 23.Kensler T.W., Wakabayashi N., Biswal S. Cell survival responses to environmental stresses via the keap1-nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J., Wang H., Shen R., Fang J., Yang Y., Dai W., Zhu Y., Zhou M. Mitochondrial-targeted antioxidant mitoq provides neuroprotection and reduces neuronal apoptosis in experimental traumatic brain injury possibly via the Nrf2-ARE pathway. Am. J. Transl. Res. 2018;10:1887–1899. [PMC free article] [PubMed] [Google Scholar]
- 25.Ouyang W., Liu C., Pan Y., Han Y., Yang L., Xia J., Xu F. SHP2 deficiency promotes Staphylococcus aureus pneumonia following influenza infection. Cell Prolif. 2020;53 doi: 10.1111/cpr.12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouyang W., Zhou H., Liu C., Wang S., Han Y., Xia J., Xu F. 25-Hydroxycholesterol protects against acute lung injury via targeting MD-2. J. Cell Mol. Med. 2018;22:5494–5503. doi: 10.1111/jcmm.13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J., Huang J., Qi T., Huang Y., Lu Y., Zhan T., Gong H., Zhu Z., Shi Y., Zhou J., Yu L., Zhang X., Cheng H., Ke Y. SHP2 protects endothelial cell barrier through suppressing VE‐cadherin internalization regulated by MET‐ARF1. FASEB. J. 2019;33:1124–1137. doi: 10.1096/fj.201800284R. [DOI] [PubMed] [Google Scholar]
- 28.Cen M., Yao Y., Cui L., Yang G., Lu G., Fang L., Bao Z., Zhou J. Honokiol induces apoptosis of lung squamous cell carcinoma by targeting FGF2-FGFR1 autocrine loop. Cancer Med. 2018;7:6205–6218. doi: 10.1002/cam4.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shoshan-Barmatz V., Krelin Y., Shteinfer-Kuzmine A. VDAC1 functions in Ca2+ homeostasis and cell life and death in health and disease. Cell Calcium. 2018;69:81–100. doi: 10.1016/j.ceca.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Watson E.C., Grant Z.L., Coultas L. Endothelial cell apoptosis in angiogenesis and vessel regression. Cell Mol. Life Sci. 2017;74:4387–4403. doi: 10.1007/s00018-017-2577-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar P., Shen Q., Pivetti C.D., Lee E.S., Wu M.H., Yuan S.Y. Molecular mechanisms of endothelial hyperpermeability: implications in inflammation. Expert Rev. Mol. Med. 2009;11 doi: 10.1017/S1462399409001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu H., Poirier C., Cook T., Traktuev D.O., Merfeld-Clauss S., Lease B., Petrache I., March K.L., Bogatcheva N.V. Conditioned media from adipose stromal cells limit lipopolysaccharide-induced lung injury, endothelial hyperpermeability and apoptosis. J. Transl. Med. 2015;13 doi: 10.1186/s12967-015-0422-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Y., Lv N., Lu Z., Zheng Y.Y., Zhang W.C., Chen C., Peng Y.J., He W.Q., Meng F.Q., Zhu M.S., Chen H.Q. Deletion of myosin light chain kinase in endothelial cells has a minor effect on the lipopolysaccharide-induced increase in microvascular endothelium permeability in mice. FEBS J. 2012;279:1485–1494. doi: 10.1111/j.1742-4658.2012.08541.x. [DOI] [PubMed] [Google Scholar]
- 34.Friedman J.R., Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornmann B. The endoplasmic reticulum-mitochondria encounter structure: coordinating lipid metabolism across membranes. Biol. Chem. 2020;401:811–820. doi: 10.1515/hsz-2020-0102. [DOI] [PubMed] [Google Scholar]
- 36.Islam M.N., Das S.R., Emin M.T., Wei M., Sun L., Westphalen K., Rowlands D.J., Quadri S.K., Bhattacharya S., Bhattacharya J. Mitochondrial transfer from bone-marrow–derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat. Med. 2012;18:759–765. doi: 10.1038/nm.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy M.P. Development of lipophilic cations as therapies for disorders due to mitochondrial dysfunction. Expert Opin. Biol. Ther. 2001;1:753–764. doi: 10.1517/14712598.1.5.753. [DOI] [PubMed] [Google Scholar]
- 38.Murphy M.P., Smith R.A. Targeting antioxidants to mitochondria by conjugation to lipophilic cations. Annu. Rev. Pharmacol. Toxicol. 2007;47:629–656. doi: 10.1146/annurev.pharmtox.47.120505.105110. [DOI] [PubMed] [Google Scholar]
- 39.Chen S., Wang Y., Zhang H., Chen R., Lv F., Li Z., Jiang T., Lin D., Zhang H., Yang L., Kong X. The antioxidant MitoQ protects against CSE-induced endothelial barrier injury and inflammation by inhibiting ROS and autophagy in human umbilical vein endothelial cells. Int. J. Biol. Sci. 2019;15:1440–1451. doi: 10.7150/ijbs.30193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu Q., Ren J., Li G., Wu J., Wu X., Wang G., Gu G., Ren H., Hong Z., Li J. The mitochondrially targeted antioxidant MitoQ protects the intestinal barrier by ameliorating mitochondrial DNA damage via the Nrf2/ARE signaling pathway. Cell Death Dis. 2018;9 doi: 10.1038/s41419-018-0436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li R., Ren T., Zeng J. Mitochondrial coenzyme Q protects sepsis-induced acute lung injury by activating PI3K/Akt/GSK-3 β/mTOR pathway in rats. BioMed. Res. Int. 2019;2019 doi: 10.1155/2019/5240898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peake B.F., Nicholson C.K., Lambert J.P., Hood R.L., Amin H., Amin S., Calvert J.W. Hydrogen sulfide preconditions the db/db diabetic mouse heart against ischemia-reperfusion injury by activating Nrf2 signaling in an Erk-dependent manner. Am. J. Physiol. Heart Circ. Physiol. 2013;304:1215–1224. doi: 10.1152/ajpheart.00796.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsushima M., Liu J., Hirao W., Yamazaki H., Tomita H., Itoh K. Emerging evidence for crosstalk between Nrf2 and mitochondria in physiological homeostasis and in heart disease. Arch. Pharm. Res. 2020;43:286–296. doi: 10.1007/s12272-019-01188-z. [DOI] [PubMed] [Google Scholar]
- 44.Khan A.U.H., Allende-Vega N., Gitenay D., Garaude J., Vo D.N., Belkhala S., Gerbal-Chaloin S., Gondeau C., Daujat-Chavanieu M., Delettre C., Orecchioni S., Talarico G., Bertolini F., Anel A., Cuezva J.M., Enriquez J.A., Cartron G., Lecellier C.H., Hernandez J., Villalba M. Mitochondrial Complex I activity signals antioxidant response through ERK5. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-23884-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starkov A.A. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann. N. Y. Acad. Sci. 2008;1147:37–52. doi: 10.1196/annals.1427.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robb E.L., Gawel J.M., Aksentijević D., Cochemé H.M., Stewart T.S., Shchepinova M.M., Qiang H., Prime T.A., Bright T.P., James A.M., Shattock M.J., Senn H.M., Hartley R.C., Murphy M.P. Selective superoxide generation within mitochondria by the targeted redox cycler MitoParaquat. Free Radic. Biol. Med. 2015;89:883–894. doi: 10.1016/j.freeradbiomed.2015.08.021. [DOI] [PubMed] [Google Scholar]
- 47.Wang P., Geng J., Gao J., Zhao H., Li J., Shi Y., Yang B., Xiao C., Linghu Y., Sun X., Chen X., Hong L., Qin F., Li X., Yu J.S., You H., Yuan Z., Zhou D., Johnson R.L., Chen L. Macrophage achieves self-protection against oxidative stress-induced ageing through the Mst-Nrf2 axis. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-08680-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slocum S.L., Kensler T.W. Nrf2: control of sensitivity to carcinogens. Arch. Toxicol. 2011;85:273–284. doi: 10.1007/s00204-011-0675-4. [DOI] [PubMed] [Google Scholar]
- 49.Moritani C., Kawakami K., Shimoda H., Hatanaka T., Suzaki E., Tsuboi S. Protective effects of rice peptide oryza peptide-P60 against oxidative injury through activation of Nrf2 signaling pathway in vitro and in vivo. ACS. Omega. 2020;5:13096–13107. doi: 10.1021/acsomega.0c01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.