Abstract
Circulation of cerebrospinal fluid (CSF) plays an important role during development. In zebrafish embryo, the flow of CSF has been found to be bidirectional in the central canal of the spinal cord. In order to compare conditions and genetic mutants across each other, we recently automated the quantification of the velocity profile of exogenous fluorescent particles in the CSF. We demonstrated that the beating of motile and tilted cilia localized on the ventral side of the central canal was sufficient to generate locally such bidirectionality. Our approach can easily be extended to characterize CSF flow in various genetic mutants. We provide here a detailed protocol and a user interface program to quantify CSF dynamics. In order to interpret potential changes in CSF flow profiles, we provide additional tools to measure the central canal diameter, characterize cilia dynamics and compare experimental data with our theoretical model in order to estimate the impact of cilia in generating a volume force in the central canal. Our approach can also be of use for measuring particle velocity in vivo and modeling flow in diverse biological solutions.
Keywords: Cerebrospinal fluid, Fluid mechanics, Central canal, Bidirectionality, Flow velocity profile, Cilia beating, Zebrafish, Development
Background
In zebrafish embryos, cerebrospinal fluid (CSF) flow is observable from 24 h post fertilization (hpf) in the central canal of the spinal cord ( Sternberg et al., 2018 ; Cantaut-Belarif et al., 2018) and later extends to the brain ventricles ( Olstad et al., 2019 ). In the embryonic central canal, CSF flows bidirectionally: towards the tail on the ventral side and towards the head on the dorsal side ( Sternberg et al., 2018 ; Cantaut-Belarif et al., 2018; Thouvenin et al., 2020 ). This bidirectionality is caused by the beating of polarized motile cilia mostly active in the ventral region of the central canal driving a caudally directed movement of fluid in the ventral region and creating a counter flow in the dorsal region.
Because of the narrow geometry of the central canal and the large contribution of Brownian motion, classical analysis tools failed to quantify the bidirectional properties of CSF flow. Recently, we developed an automated method based on kymograph analysis allowing the quantification of embryonic CSF flow ( Thouvenin et al., 2020 ). One challenge now is to generalize this approach in order to compare a variety of genetic animal models and experimental conditions. This is of special interest for investigations on cilia-defective mutants in which motility defects are partial and the consequences on flow are not fully understood.
The goal of this protocol is to guide the computation of CSF flow profiles from fluorescence measurements. We developed a user-friendly interface program to generate flow profiles from collected data. We additionally provide a protocol to compare experimentally measured profiles of embryonic CSF flow to a theoretical profile. Our theoretical model relies on the assumption that the average flow rate is null. In this case, the volume force that gives rise to CSF flow can be computed and compared between different conditions. The volume force depends on different cilia parameters:, where is the average cilia beating frequency, the width of the region occupied by the cilia, the viscosity, and a dimensionless parameter. We finally show how to quantify the main cilia frequency using transgenic embryos with cilia labeled by fluorescent proteins.
Materials and Reagents
2 ml Eppendorf tubes (Eppendorf, catalog number: 0030120094)
Glass bottom Petri dish (MatTek Corporation, catalog number: P50G-1.5-14-F)
Home-made Microinjection needles from Borosilicate Glass with Filament, OD 1 mm, ID 0.5 mm, length 10 cm, Fire polished (Sutter Instrument Company, catalog number: BF100-50-10)
-
FluoSphereTM size kit #2, six sizes (Molecular Probes, catalog number: F8888)
Note: This protocol is optimized for 20 nm carboxylate FluoSpheres of center wavelength 505/515 nm (yellow/green).
Nacre, AB and Tüpfel long fin (TL) strains of Danio rerio aged between 1 and 6 days post fertilization (dpf) can be used to achieve the CSF flow profile quantification. When optical imaging is performed at the embryonic stage, the pigmentation observed in AB and TL lines is not affecting either imaging using spinning disk at shallow depth nor analysis. The comparison of experimental measures of CSF flow profile with the theoretical model only holds for zebrafish embryos. We used the Tg(β-actin:Arl13b-GFP) transgenic line to image cilia in vivo ( Borovina et al., 2010 )
3,000 MW TexasRed dextran, Excitation/Emission wavelength: 595/615 (ThermoFisher Scientific, catalog number: D3329)
-
α-bungarotoxin (TOCRIS, catalog number: 2133)
Note: We injected α-bungarotoxin solution to paralyze the fish, see Recipes to prepare the solution.
Tricaine (ethyl 3-aminobenzoate methane sulfonate salt, Sigma-Aldrich, catalog number: A5040)
UltraPureTM low melting point agarose (Invitrogen, catalog number: 16520100)
NaCl (Sigma-Aldrich, catalog number: S7653)
KCl (Sigma-Aldrich, catalog number: P9333)
HEPES (Sigma-Aldrich, catalog number: H3375)
Alpha-D-Glucose (Sigma-Aldrich, catalog number: 158968)
CaCl2 dihydrate (Sigma-Aldrich, catalog number: H3375)
NaOH (Sigma-Aldrich, catalog number: 71687)
Artificial Cerebrospinal fluid (aCSF) (see Recipes)
α-bungarotoxin solution (see Recipes)
TexasRed solution (see Recipes)
Injection Mix 1 (see Recipes)
Injection Mix 2 (see Recipes)
Equipment
Fine forceps (Fine Science Tools, catalog number: 11412-11)
Spinning disk microscope (Leica Microsystems, model: Inverted Leica DMI8) equipped with a Hamamatsu Orca Flash 4.0 camera (Maximum 100 frames per second full frame), using a 40× water immersion objective (N.A. = 0.8) or any similar
Pipette puller (Narishige Ltd., model: PC-10 or PC-100 puller, or any similar)
Sonicator (VWR, Ultrasonic Cell Disruptor, Vibra-CellTM VCX 130)
Picospitzer injector (World Precision Instruments, Sarasota, USA)
Software
MATLAB 2018b (MathWorks, Mountain View, CA, USA) and image processing toolbox
Fiji ( Schindelin et al., 2012 )
Microsoft Excel 2010 (for .xlsx output format)
Procedure
-
Injections and imaging of fluorescent beads
Notes:
To measure CSF flow profiles in the central canal of the spinal cord, we inject in brain ventricles a solution of 20 nm fluorescent beads diluted in artificial CSF (aCSF). Prior to injection, the sonication of this solution is a critical step to avoid aggregates of beads that would not enter the central canal. We successfully measured flow with beads of 20 nm and 45 nm but when diameter reaches 100 nm, beads fail to enter the central canal even after sonication. The bigger the particles, the brighter they are, and the less Brownian motion they experience, making the flow quantification more precise, but only if the beads can reach the central canal. Injection in the caudal central canal (Thouvenin et al., 2020) is possible but more complicated and not recommended as it may alter the CSF flow in the rostral central canal.
We recommend injecting beads together with TexasRed to make sure that the injection was successful and to identify the boundaries of the central canal if needed. If so, 0.2% TexasRed can be added to the injection mix after the sonication step (injection Mix 2).
From our experience, injections in the brain ventricles are easier (especially at larval stage) if the fish is dorsally-mounted. It is possible to perform the injection on dorsally-mounted animals, and then flip the agar block or unmount the fish to mount it again laterally in a new dish. As mounting/unmounting can increase the risk of hurting the larva, flipping a block of agar cut with a fine surgical knife (Fine Science Tools, catalog number: 10073-14) is recommended.
To avoid motion artifacts, imaging of beads trajectory has to be performed on paralyzed animals. We simply added α-bungarotoxin directly to the injection mix in order to inject the animal only once and reduce experiment time.
Video 1 illustrates the experimental procedure starting from the embryo dechorionation and mounting, and shows the ventricular injection.
-
Injection mix and injection pipette preparation
Dilute fluorescent beads (FluoSpheres) to a 2% concentration (v/v) in aCSF in a final volume of at least 100 µl. We also recommend using large 2 ml microtubes in order to avoid dispersion of the solution during the sonication step. Keep the mix on ice.
Sonicate beads by giving 3 pulses of 3-4 s each at a power output of 50 W. Make sure the mix containing diluted beads stays on ice (sonication is exothermic).
Pull microinjection needles from borosilicate glass capillaries with a 2-step needle puller. Adjust temperature and pulling force to produce a long and sharp funnel shape needle with an approximate tip diameter of 1-3 µm (equivalent to egg injection pipettes).
Prepare injection Mix 1 or Mix 2 (see Recipes).
Fill the pipette with 4 μl of injection Mix.
Connect the pipette to the Picospritzer device (Parker, Hollis, NH, USA).
Cut the extremity of the pipette with pliers to get a 1-3 nanoliters injection drop. The volume of the drops can be calibrated prior to the experiment.
(Optional) To calibrate the volume of the drops at a fixed ejection pressure, fill the injection pipette with a defined and fixed volume of aCSF (e.g., 0.2 µl), and connect the pipette to the picospitzer device. Prepare a dish containing mineral oil and inject drops in the mineral oil until the injection pipette is empty. Count the number of drops ejected and use it to calculate the average volume of each drop. The volumes of the drops can be adjusted by changing the ejection pressure or the diameter of the pipette tip.
-
Embryos mounting and injection (see Video 1)
Dechorionate 26-28 hpf embryos manually using fine forceps. Other dechorionation techniques such as the enzymatic digestion should be equally efficient.
Anesthetize the embryos in 0.02% w/v Tricaine solution (or in a dish in contact with ice) to stop spontaneous twitches during the mounting step and rinse once mounting is done.
Mount the embryos laterally in 1.5% w/v low melting point agarose. Make sure to orient thoroughly the posterior axis of the animal to image sufficiently flat and long portions of the lumen of the central canal. Add at least 1 ml of system water when agarose has solidified so that the embryo can breathe and to dilute the remaining tricaine solution.
Inject previously prepared solution mix in the hindbrain (or ‘rhombencephalic’) ventricle (Figure 1). If 0.2% TexasRed were added (injection Mix 2), injection quality can be controlled by observing the brain ventricles filled with the colored solution. If not (injection Mix 1), injection quality can be assessed by observing a slight and transient swelling of the ventricle (see Video1).
Put injected embryos at 28 °C for 1 h to allow the diffusion of fluorescent beads down to the central canal. Make sure system water covers the entire surface of the dish to compensate for evaporation.
-
Optical imaging of fluorescent beads in the central canal
Notes:
Time-lapse images can be acquired at 26 °C using a spinning disk confocal microscope mounted in a thermostatic chamber. This allows a better control of temperature variations that may affect CSF flow properties and/or cilia beating properties.
We recommend to orient the embryo always in the same direction with the rostral side on the left and the dorsal side on the top. If not, correct orientation can be restored via image processing. Note that, in wild type embryos, the ventral wall of the central canal is typically flatter than the dorsal wall.
Imaging could be performed with any imaging system as long as the signal to noise ratio (SNR) is high enough, and the imaging speed is above a few frames/s. Upright or inverted spinning disk can be chosen. Spinning disk imaging seems the most adequate but widefield microscope could be suitable as well with bright fluorescent beads. Confocal or two-photon microscopes could also be used, although the imaging speed might be limited with classical confocal setups and in two-photon microscopes, the salt-and-pepper noise generated might be problematic.
If TexasRed is injected together with 505/515 nm emitting beads, make sure that the emission filters of the microscope allow dual color imaging (avoid using long-pass filters that would lead to bleed-through).
Because imaging was mostly performed in the sagittal plane at the embryonic and larval stage using visible laser for excitation, pigmentation was not an issue and did not therefore require the use of PTU.
As the central canal shape and cilia properties may differ along the rostrocaudal axis, we recommend imaging always at the same rostro-caudal position. In our case, we focused on 3 segments above the yolk extension (Figure 1). Because spinning disk microscopes perform sharp optical sectioning, we advise using either the Differential Interference Contrast (DIC) or transmitted light channel to image somites and central canal together. If TexasRed was injected, the red epifluorescence channel can be superimposed to visualize the central canal using epifluorescence before switching to the spinning disk mode (Figure 1).
Perform time-lapse 2D acquisitions at 10 frames/s for 30 s. Carefully mount the fish so that the central canal is horizontal over a long distance. By choosing the Z plane in which the apparent diameter of the central canal is maximal in the sagittal plane, imaging occurs at the center of the central canal, onto the midline. By doing so, with the 40× NA = 0.8 objective, the central canal mostly fits within the optical section.
FluoSpheres flowing in the CSF are photostable: high laser power can be used with minimal photobleaching in order to get high signal to noise ratio.
Use the program GeneProfile to generate the CSF flow profile from the acquired data. See Data analysis section for more details.
Fit the CSF flow profile to the theoretical bidirectional flow expected in wild type embryos using the same program GeneProfile. See Data analysis section for more details.
-
Quantification of the kinematics of motile cilia
-
Embryo mounting and paralysis
Dechorionate 26-28 hpf Tg(β-actin:Arl13b-GFP) embryos ( Borovina et al., 2010 ) screened for GFP.
Put the embryos in 0.02% w/v Tricaine solution (or in a dish in contact with ice) to stop spontaneous twitches during the mounting step.
Mount the embryos laterally in 1.5 % w/v low melting point agarose in a glass bottom Petri dish. When agarose has solidified, add system water. Make sure that Tricaine is diluted out.
Paralyze embryos. Prepare injection pipette similarly to Step A1. Fill the pipette with a solution of 500 µM α-bungarotoxin. Perform a single 1 nl injection in the caudal-most muscles and check that it induces a full paralysis. If the central canal needs to be visualized properly, TexasRed can be injected together with α-bungarotoxin in the muscles, by replacing aCSF with TexasRed solution when diluting the α-bungarotoxin stock aliquots (see Recipes). TexasRed is able to cross the blood brain barrier and penetrate inside the central canal at early developmental stages.
-
Optical imaging of cilia dynamics
Make sure the temperature of the room is controlled at 26 °C ± 2 °C to avoid variations in cilia beating frequency across experiments.
Using a spinning disk microscope, monitor the motility of cilia via time-lapse acquisitions in 2D using a blue laser for excitation of the Arl13-GFP. Acquire a single image of TexasRed injected in the brain ventricles using a green laser for excitation to reliably measure the central canal diameter. In order to capture the highest beating frequencies, acquisition rates of 100-200 Hz should be chosen. If the system relies on a CMOS camera, the frame rate can be increased by positioning the central canal along the lines of the camera and reducing to the minimum the number of lines to image the central canal. Exposure time should be sufficient to visualize GFP-positive cilia.
Use the program cilia analysis to extract cilia beating frequency, length and angle. See Data analysis section for more details.
-
Video 1. Experimental procedure for mounting and injecting fluorescent beads in zebrafish embryos.
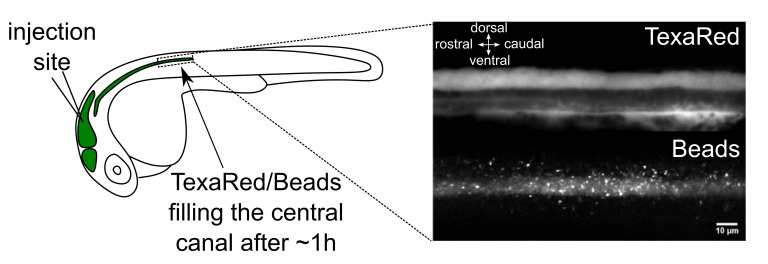
Figure 1. Brain ventricle injection site and injection quality assessment.
Left panel: Injection site of fluorescent beads in the hindbrain ventricle of a 30 h post fertilization (hpf) embryo. An hour after injections in the hindbrain ventricle, 20 nm beads have propagated along the central canal. Right panel: TexasRed (top) and 20 nm beads (bottom) imaging with spinning disk in the central canal of the spinal cord above the yolk extension. Scale bar: 10 µm.
Data analysis
On top of the experimental procedures, we detail below two independent analysis workflows.
The first analysis (Section A) enables to obtain the CSF flow profile from the time series of bead trajectories acquired in Part Procedure-A. This also allows measuring the total CSF flow rate (Section B), which is expected to be null in WT embryos ( Thouvenin et al., 2020 ). If adequate (see conditions below), the experimentally measured CSF flow can be fitted to a bidirectional flow model ( Thouvenin et al., 2020 ) in order to extract the volume force generated by the motile cilia.
The second analysis workflow (Section C) uses the cilia beating movies (Procedure-B) to extract cilia parameters, including each cilia main beating frequency, length, and angle.
If appropriate, the last section (Section D) aims to combine the outputs from the two analysis workflows and extract a parameter we called α, an ad hoc coupling parameter that measures how multiple cilia efficiently work together to generate a flow.
-
CSF flow profile generation
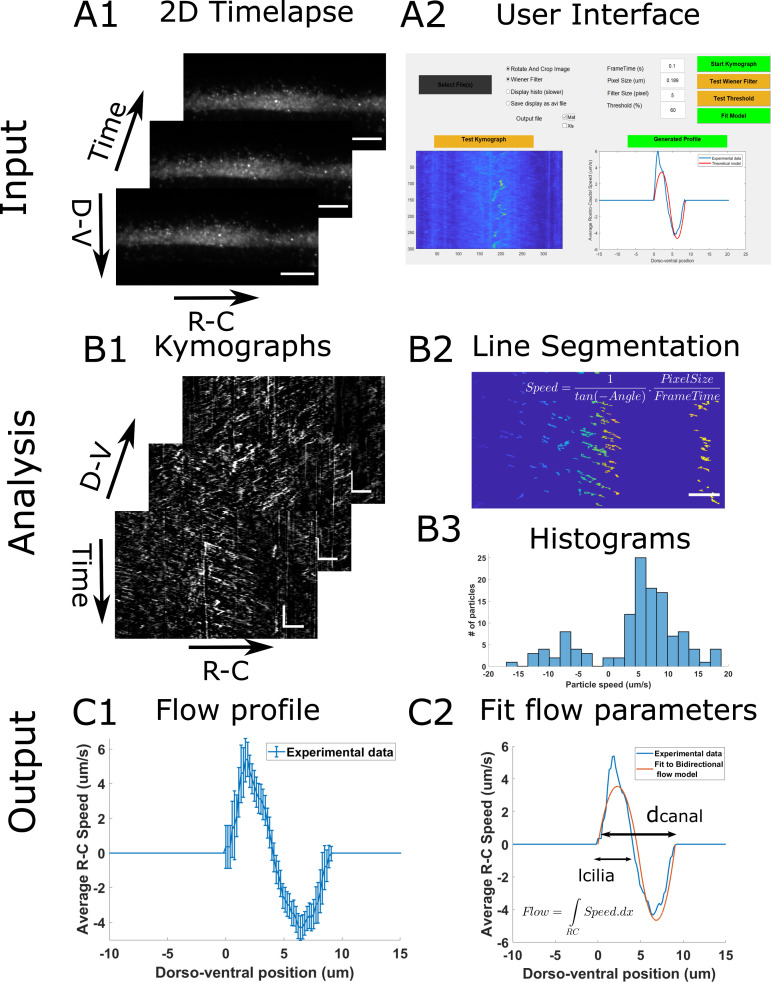
Specifically for this protocol, we developed a user-interface platform to allow users to generate CSF flow profiles as easily as possible. Here, we present the analysis workflow (Figure 2) and how to generate a first CSF flow profile from the fluorescent beads measurements. More subtle fine tuning of parameters is available within the user interface to adapt to variable imaging conditions, and is fully described in the document ManualGeneProfile.pdf file that can be found with the software.
As input, the analysis takes 2D time lapses of beads flowing in the central canal (Figure 2A1). In order to generate kimographs, we swap for a given dorsoventral position the axes so that the X axis corresponds to the rostrocaudal position and the Y axis to time. Then, the beads trajectories appear as lines whose slopes reflect the direction and speed of the particles along the rostrocaudal axis. In order to build the flow profile, the program filters each kymograph and performs automatic segmentation of all lines in each kymograph (Figure 2B2). It then extracts the slope of each line, and converts it into the particle velocity, in order to build a histogram of velocities for each dorso-ventral position (Figure 2B3). By calculating the average velocity at each position, we generate the CSF flow profile (Figure 2C1).
-
Launch program
Run Main_GeneProfile.m (requires MATLAB 2018b or later).
Alternatively, download and install the standalone application. Once it is installed, go to the command window and navigate to the installed folder. Run: application\GeneProfile.
The user interface window in Figure 2A2 opens.
Select .tif files to analyze. Multiple files can be selected at once, and they will be processed one by one.
Enter the frame time in seconds and pixel size in microns, corresponding to the experimental parameters.
For the analysis to work, the central canal should be as horizontal and flat as possible. Choose the longest horizontal portion of the canal with a few pixels outside the central canal on both sides (Figure 2A1). The rotation and crop of the region of interest can be done either manually, with Fiji, or any equivalent software, or using the provided user interface by clicking the RotationAndCrop button. In this case, a new interface aiming to select a region of interest appears when the StartKymograph button is pressed.
Run Start Kymograph to calculate the kymograph and the histograms of beads velocity at each dorso-ventral position. It then measures the mean of each histogram to compute the flow profile. In case multiples input .tif files are used, each profile is displayed sequentially. The data is saved in the Matlab workspace in a matrix called CSFProfile, and is also saved in the desired output.
Possible outputs are .mat, .xlsx, or both, and set in the user interface. For the .xlsx output, Excel has to be installed on the computer. The .mat generates a structure that contains, for each file, the dorso-ventral position, the flow at each position, the standard error, as well as the speed histograms at the extrema of the flow profile. In the .xlsx output, the data from each file is saved in a different sheet, and the dorso-ventral position, the flow at each position, and the standard error are saved.
The program ends by closing the interface window.
-
-
Fitting the velocity profile with a model
In Thouvenin et al. (2020) , we developed a simple model accounting for the bidirectionality of CSF flow in zebrafish embryos. In this protocol, the users can use our script to automatically fit their experimental data of CSF flow to the theoretical model we developed (Figure 2C2). This allows users to check 1) whether their experimental conditions lead to a similar CSF flow than the one we observed in the central canal of wild type (WT) zebrafish embryos, and if this is the case 2) to extract quantitative parameters from the fit. The program allows extracting 4 hydrodynamics parameters: 1) The volume force generated by motile cilia, 2) the pressure gradient that is established in the canal to oppose the cilia beating, 3) the width of the region bearing motile cilia, and 4) the diameter of the central canal. In order for the fit to be meaningful, the two assumptions of our bidirectional flow model should be respected: a cylindrical geometry for the central canal and the “no net flux” condition ( Thouvenin et al., 2020 ). As a reminder, under these two assumptions, we showed that the averaged velocity profile can be fairly described by a piecewise second-order polynomial, defined as:
where ,, , and are expressed as a function of the parameters of the problem:
,
,Here is the diameter of the channel and the viscosity of the CSF, is the pressure gradient and is the average force per unit volume generated by the cilia. The latter two parameters can be measured experimentally from the CSF velocity profiles processed with the GeneProfile interface.
Our theoretical model describes a symmetric bidirectional flow for which the net total flow rate is null. In the user interface, before launching the fitting tool, we provide the user an estimate of the bidirectionality of the flow, called that is defined as:
varies between 0% for a purely monodirectional flow and 100% for a purely bidirectional flow (the flow rate advected caudally equals the flow rate advected rostrally).
We advise users not to perform the fit of velocity profiles for values of < 70%, because under this arbitrary threshold, the “no flux” condition is no longer valid, and therefore the parameters of the fit are meaningless.
Once the flow profile is calculated (Data analysis Section A), the “Fit Model” button on the right (Figure 2A2) turns green. The fit can be performed by pushing this button.
-
Two possibilities can arise:
If < 70%, we estimate that the flow is not bidirectional enough to fit the velocity with our model, and display the warning message “We advise the user not to go further”. If the two assumptions for our model are not respected, we advise to click on the “Stop here” button in order to stop the fitting process. If the measured flow profile was robustly measured, having a low β means that the flow could not be simply explained by the action of motile cilia in a closed cylindrical geometry and that another model should be developed by taking into account other physical effects.
If > 70%, the flow can be reasonably fitted with the simple model, and a verification message is sent to the user. Click on “Yes”.
The experimental velocity profile (in blue on the bottom right plot of the user interface, Figure 2A2) is then fitted by the theoretical model (in red on the plot). The plot is also saved in .fig and .png format (Figure 2C2). The .fig file allows the user to modify the plot, and do aesthetic changes, as well as to save in vector formats (.eps, .pdf, .svg).
-
Several information can be extracted from the fit, which are detailed below. The data are stored in either an .xls file, and/or a .mat file, called “TheoreticalProfiles”, where:
The velocity is stored in a 10000 x 1 vector called “Profile” (in μm/s) and the rostro-caudal position in a 10000 x 1 vector called “X” (in μm).
The volume force generated by the cilia is stored in the variable called “f_v” (in N.m-3).
The pressure gradient , opposing the cilia beating due to the closed geometry of the central canal, is stored in the variable “GradP” (in N.m-3).
The width of the ciliary region, where the volume force is generated in the model, is stored in the variable “l_cilia” (in μm).
The measured diameter of the central canal is stored in the variable “Diameter” (in μm). Note that, if required, we advise to use a direct measurement with TexasRed to measure the central canal diameter, since deducing the diameter from the kymograph may be imprecise.
-
Cilia frequency measurement
This section aims to describe the analysis protocol to estimate the main beating frequency of cilia (see Thouvenin et al., 2020 ), from the fluorescence cilia time lapse acquisitions described in the section Procedure B.
Similarly to section Data Analysis A, we describe here the principle of the analysis workflow (Figure 3), as well as key instructions to perform a first analysis. Detailed instructions, as well as descriptions of fine-tuning parameters are available in an external document ManualCilia.pdf that can be found with the shared code.
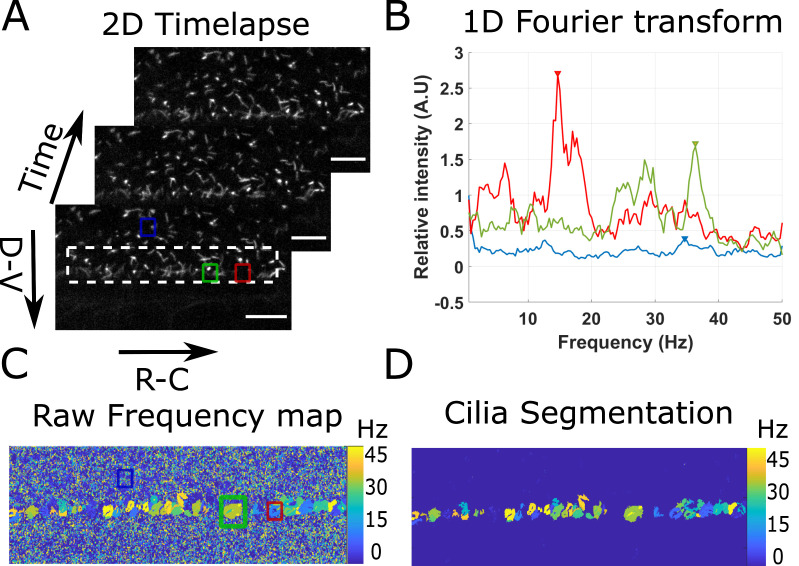
The program first loads the imaging data with cilia dynamics versus time (Figure 3A), and applies a local average filter (of size 4 by default) to increase the cilia SNR. For each pixel in the filtered data, the time series is extracted and Fourier transformed (Figure 3B). The 5 maximal peaks of the Fourier spectrum are extracted, but, by default, only the first one is used. The frequency of the other peaks can be used for validation (e.g., if sampling errors are made, the sum of the frequencies of the first and second peak is equal to the acquisition frequency). A 2D image with the main frequency found at each pixel is thus created (Figure 3C). In noisy regions, it outputs a random frequency, but in cilia regions it draws regions of interests of a given frequency that we considered to be single cilium. Each of these regions of interest containing more than 40 pixels (7.5 μm2) is finally segmented and analyzed. The parameters frequency, diameter, eccentricity, area, angle, and major axis length are extracted and associated to their corresponding cilia parameters.
If a comparison between dorsal and ventral cilia is of interest ( Thouvenin et al., 2020 ), the program allows to manually draw a line at the center of the central canal and classify cilia as dorsal or ventral with respect to their relative position from the central line. This procedure is not described further here, but can be found in the document “ManualCilia.pdf” located in the same folder as the shared Matlab code.
Open the program NewAnalysisCilia.m with MATLAB.
If the acquisition frequency is different from 100 Hz, correct the value at the beginning of the code (Line 8).
Run the code.
Select a folder where the .tif files with cilia imaging data can be found. If several acquisitions are located in the same folder, all .tif files will be analyzed sequentially, and cilia parameters from all experiments will be concatenated.
All the rest of the analysis is automatic afterwards according to the workflow described above.
The programs outputs one .mat file with 4 matrices. The most interesting matrix is called AllFreq and contains 9 parameters (lines) for all detected cilia (columns). If several files are analyzed, the matrix AllFreq concatenates the parameters of all cilia from all files and saves the number of the file from which the cilia was taken. The 6 first parameters are respectively the beating frequency, the apparent diameter of the region described by the cilia, its angle, eccentricity, area, and major axis. The seventh parameter is an indicator for ventral (1) or dorsal (0) cilia if a central line was drawn (-1 otherwise). The eighth parameter is the number of the file from which the cilia was measured. The last parameter is an indicator of proximity to the central canal (1 if close to the central canal) if a line at the center of the central canal is drawn, in order to exclude cilia outside the central canal. The Freq and Freq_FromPSD matrices display the 5 main frequencies found at each pixel, calculated either from a simple Fourier transform or from the power spectrum density of the last .tif file analyzed. The FrequencyMap matrix gives a 2D map with the main frequency found at each pixel after spatial filtering (Figure 3C).
-
Quantification of the volume force and cilia parameters
From the beads measurement (Procedure A), the CSF flow can be analyzed qualitatively (e.g., more or less vortices) and quantitatively by measuring the velocity profile (Data analysis A). The diameter of the central canal, as well as the flow rate were obtained from this measurement. The central canal diameter is one output of the fit of the bidirectional flow model and can be recovered because the velocity is zero on the dorsal and ventral boundaries of the central canal. The volume force generated by the collective action of cilia could also be quantified using our bidirectional flow model (Data analysis Section B). In this section, we propose to estimate the parameters characterizing cilia and the CSF that control the volume force (see definition below).
The volume force , measures the average force created by the collective action of cilia and divided by the volume of the ciliary region. It can be expressed as:
where is the viscosity of the CSF, is the average cilia frequency, is the width of the region occupied by the beating cilia and is a dimensionless parameter. For users who measured the average cilia frequency , the coefficient can be estimated, as all the other parameters are henceforth known: is the force provided by the fitting tool and can be either extracted from the fitting tool or from the cilia beating analysis. The viscosity of the CSF, can be approximated to 10-3 Pa.s, as the viscosity of water, and is supposed constant. The coupling coefficient provides a mean to compare the collective efficiency of beating cilia in order to generate a flow in either WT or mutant embryos ( Thouvenin et al., 2020 ). Note that we assumed the CSF viscosity to be a constant between wild type and mutant embryos, but this assumption should be verified.
To provide comparative data, a value of was obtained for 30 hpf wild type embryos ( Thouvenin et al., 2020 ), with μm, Hz and μ = 10-3 Pa.s, which corresponds to a volume force N/m3.
Figure 2. Principle of the CSF flow profile analysis workflow.
The CSF flow profiles are calculated using 2D time lapses of fluorescent beads flowing in the central canal (A1), and our custom user interface software (A2). The Start Kymograph button starts the analysis by first calculating kymographs from 2D time lapses by swapping dimensions (B1). Each kymograph is filtered, and all lines, corresponding to one particle trajectory, are segmented (B2). The angle of each line is then transformed to a particle velocity value, and aggregated into a speed histogram for each D-V position (B3). The flow profile is then calculated by extracting the mean and standard error of each histogram (C1). The flow profile can finally be fitted to our bidirectional flow model, to extract quantitative flow parameters (C2). Horizontal scale bars are 15 µm and vertical scale bar is 5 s.
Figure 3. Principle of the beating cilia analysis.
2D time lapses of cilia beating (A) are analyzed by measuring the time Fourier transform at each pixel (B). 3 spectra corresponding to the pixel at the center of the three regions drawn in (A) are plotted. The frequency of the peak of maximum amplitude (arrows in (B)) is extracted for all spectra, to form a frequency map (C), showing regions of constant frequency corresponding to individual cilia. All cilia are then segmented by keeping only the largest regions of constant frequency (of area above 40 pixels) (D). Scale bar is 15 µm.
Notes
From measurement on beads, CSF flow can be analyzed qualitatively to estimate recirculation spots and quantitatively to measure the velocity profile. We can extract from these measurements the diameter of the central canal as beads explore the entire lumen of the central canal as well as the flow rate.
If the overall flow rate is close to 0, the CSF flow profile can be fitted by the bidirectional flow model ( Thouvenin et al., 2020 ) to obtain the volume force generated by the collective action of cilia.
Using Tg(β-actin:Arl13b-GFP) embryos, the average cilia length and beating frequency can be extracted. Thus, if we assume the CSF viscosity to be constant, the coupling coefficient α, measuring the efficiency of the beating cilia to generate a flow, can be compared between mutants and wild type embryos.
In case error messages occur using this interface, please contact us with a precise description of the error, and we will update the code online.
The excel output may fail on some computers, especially if a "click to run" version of office is installed, or if office is not properly registered. If it is the case, please try to uninstall and reinstall office on your computer. Please contact us in case you still experience issues.
All MATLAB scripts and test files are shared on GitHub: https://github.com/wyartlab/BioProtocol_CSFflowMeasurement.
Recipes
-
aCSF solution
Prepare a solution containing in mM:
134 NaCl
2.9 KCl
1.2 MgCl2
10 HEPES
10 glucose
2 CaCl2
Note: Osmolarity has to be 290 mOsM ± 3 mOsm, and pH needs to be adjusted to 7.8 with NaOH.
-
TexasRed solution
Prepare a 0.2% solution (w/v) of TexasRed in aCSF
-
α-bungarotoxin solution
Prepare 1 μl stock aliquots of α-bungarotoxin at 1 mM
Stock solutions can be stored at -20 °C and diluted in aCSF to get working concentrations of 100 µM for brain ventricle injections, and 500 µM for muscle injections
-
Injection Mix 1
For a 10 μl final volume:
Mix 5 μl of FluoSpheres (2% v/v concentration in aCSF, sonicated)
4 μl aCSF
1 μl α-bungarotoxin (1 mM)
-
Injection Mix 2
For a 10 μl final volume:
Mix 5 μl of FluoSpheres (2% v/v concentration in aCSF, sonicated)
4 μl TexasRed (0.2% v/v concentration in aCSF)
1 μl α-bungarotoxin (1 mM)
Acknowledgments
This work was funded by Human Frontier Science Program (HFSP) Research Grant (grant n° RGP063-2018), and the New York Stem Cell Foundation (NYSCF) Robertson Investigator award (grant n° NYSCF-R-NI39) for Claire Wyart and an ICM postdoctoral fellowship kindly attributed to OT and the Big Brain Theory (BBT) program from ICM to support the salary of YCB. The research leading to these results has also received funding from the program ‘Investissements d’avenir’ ANR-10-IAIHU-06 (Big Brain Theory ICM Program), ANR-11-INBS-0011 (NeurATRIS: Translational Research Infrastructure for Biotherapies in Neurosciences).
Competing interests
The authors declare no conflict of interest.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1. Borovina A., Superina S., Voskas D. and Ciruna B.(2010). Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat Cell Biol 12(4): 407-412. [DOI] [PubMed] [Google Scholar]
- 2. Cantaut-Belarif Y., Sternberg J. R., Thouvenin O., Wyart C. and Bardet P. L.(2018). The Reissner Fiber in the Cerebrospinal Fluid Controls Morphogenesis of the Body Axis. Curr Biol 28(15): 2479-2486 e2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Olstad E. W., Ringers C., Hansen J. N., Wens A., Brandt C., Wachten D., Yaksi E. and Jurisch-Yaksi N.(2019). Ciliary Beating Compartmentalizes Cerebrospinal Fluid Flow in the Brain and Regulates Ventricular Development. Curr Biol 29(2): 229-241 e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P. and Cardona A.(2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sternberg J. R., Prendergast A. E., Brosse L., Cantaut-Belarif Y., Thouvenin O., A. Orts-Del'Immagine, Castillo L., Djenoune L., Kurisu S., McDearmid J. R., Bardet P. L., Boccara C., Okamoto H., Delmas P. and Wyart C.(2018). Pkd2l1 is required for mechanoception in cerebrospinal fluid-contacting neurons and maintenance of spine curvature. Nat Commun 9(1): 3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thouvenin O., Keiser L., Cantaut-Belarif Y., Carbo-Tano M., Verweij F., Jurisch-Yaksi N., Bardet P. L., van Niel G., Gallaire F. and Wyart C.(2020). Origin and role of the cerebrospinal fluid bidirectional flow in the central canal. Elife 9: e47699. [DOI] [PMC free article] [PubMed] [Google Scholar]