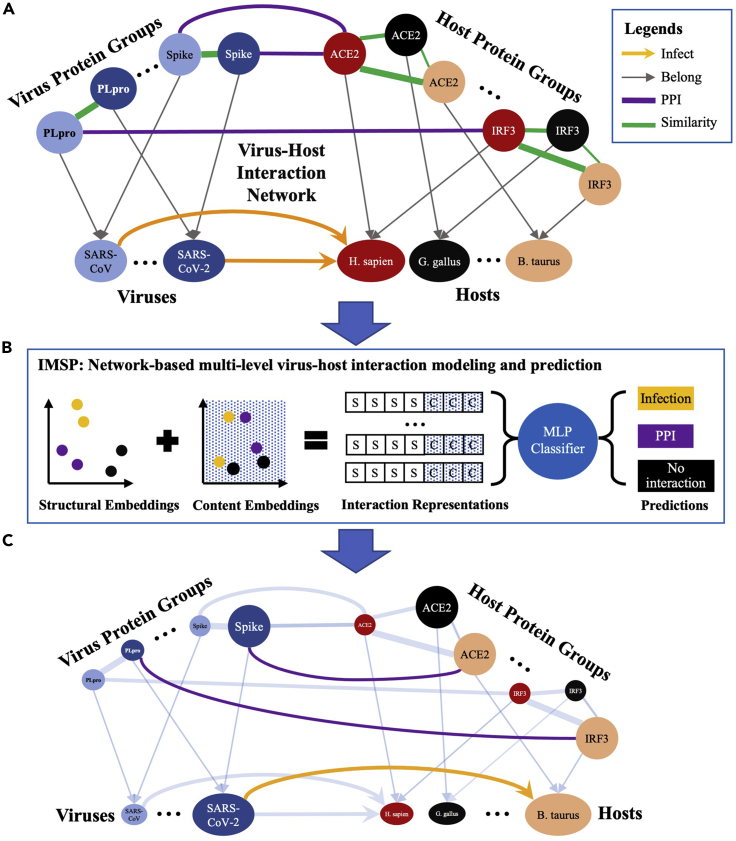
Figure 1.
Infection mechanism and spectrum prediction
(A) The virus-host interaction network. Nodes represent proteins, viruses, and hosts; edges represent relationships (i.e., PPI, infection, protein-homolog similarity, and organism-protein belonging). The color of a node indicates its organism. The thickness of a protein-homolog similarity edge indicates its level of similarity. For the full network, refer to the viral entry graph (Figure S3), interferon signaling pathway graph (Figure S4), and infection graph (Figure S5).
(B) IMSP learns a representation for each potential edge, which contains a structural embedding and a content embedding. The structural embedding captures the local structural features of an edge. The content embedding captures the attributes that reveal biological aspects of an edge. The representation of each edge is derived by concatenating its structural and content embeddings, where S stands for a structural embedding element and C stands for a content embedding element. A Multi-layer Perceptron (MLP) is trained to take the edge representations as input and reports negative (non-connected) edges whose corresponding edge representations are classified as infection or PPI. Note that no-interaction is also a potential class for the classification task. See experimental procedures for calculation of the structural and content embeddings.
(C) Exemplar predicted edges are highlighted and colored accordingly to their types. Existing edges are dimmed.