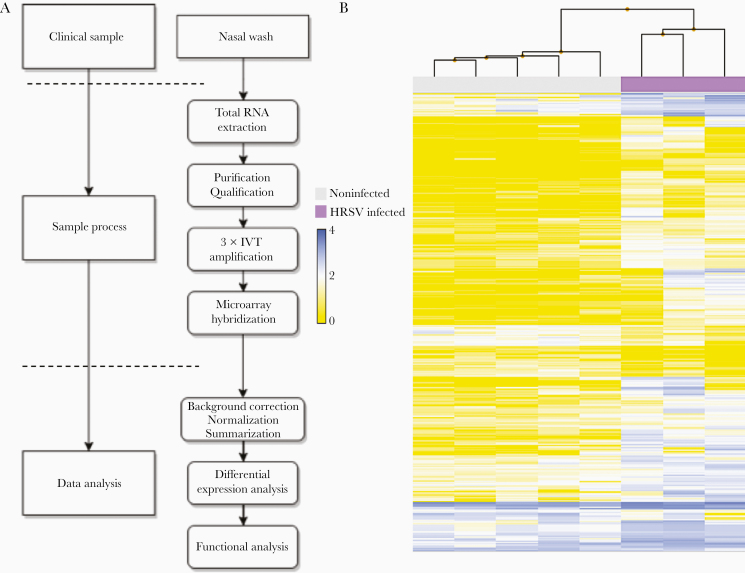
Figure 1.
Sample processing workflow and transcriptomic hierarchical clustering. A, Adapted workflow for the processing and exploitation of clinical samples with low RNA quality/quantity. B, Hierarchical clustering of the signal intensities corresponding to all infected and noninfected clinical samples evaluated in the study. The resulting clusters are representative of the degree of similarity between samples and enable clustering into infected and noninfected experimental groups. The height of the y-axis at the branching points is a measure of similarity; y-axis units are arbitrary. For representation purposes, data was autoscaled and log2 transformed. Abbreviation: IVT, in vitro transcription.