Fig. 1. Catalog of human versus Neanderthal genetic variation and the NOVA1 haplotype.
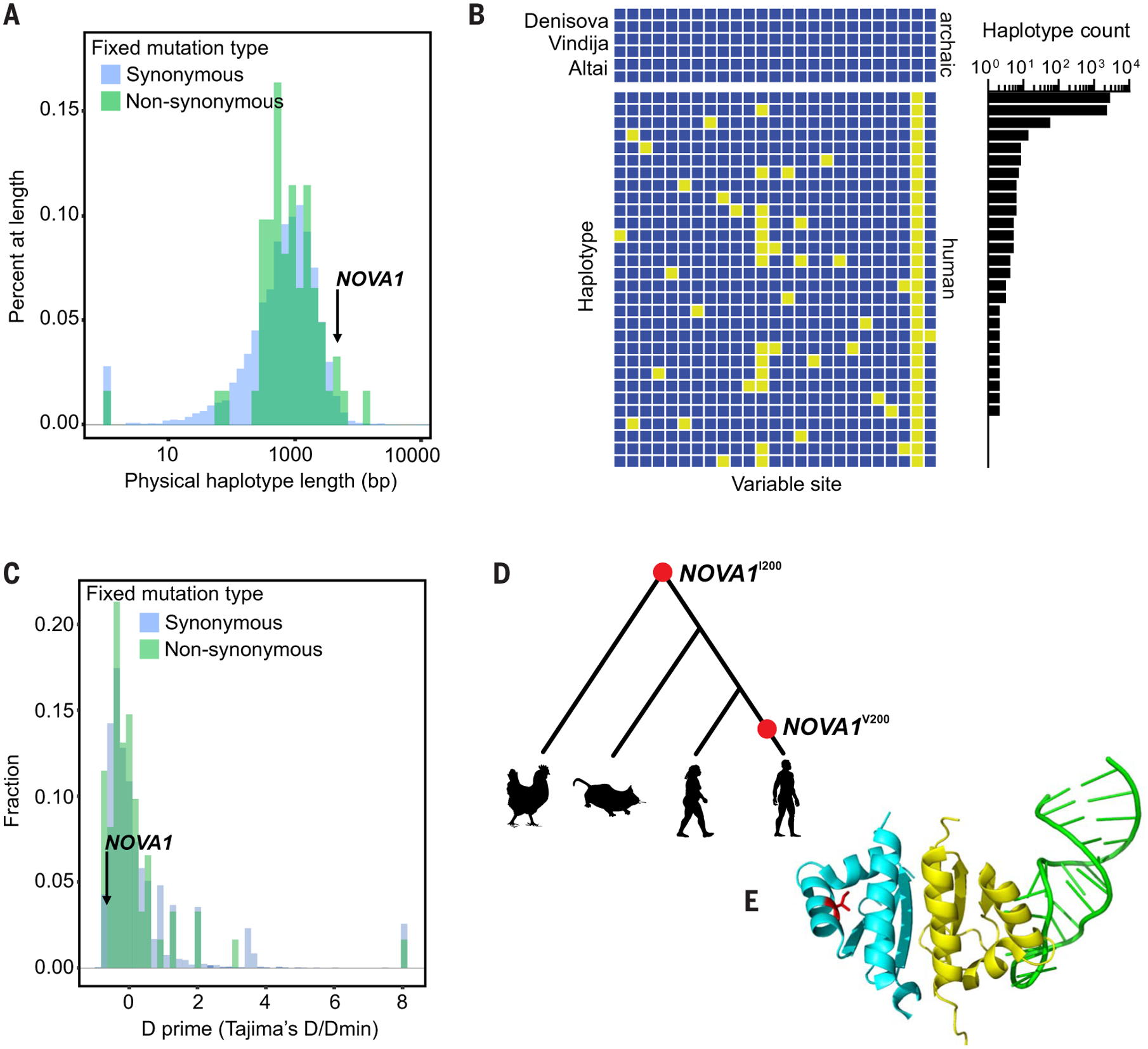
(A) Physical haplotype lengths [in base pairs (bp)] around human-specific fixed derived alleles in the 1000 Genomes Project dataset. We defined a haplotype as the distance upstream and downstream of a human-specific fixed derived allele for which no human in the 1000 Genomes Project dataset shares a derived allele with an archaic hominin. Lengths of haplotypes around both synonymous and nonsynonymous substitutions are shown. (B) Haplotypes around the human-specific fixed derived allele in NOVA1. Rows are individual haplotypes; columns are variable sites. Yellow boxes have a derived allele (different from the 1000 Genomes Project ancestral sequence). Human haplotypes are labeled by the number of (phased, haploid) human genomes that carry them. Only biallelic SNPs with reference alleles are shown, and the region is bounded by sites at which modern humans share derived alleles with archaic hominins. (C) Normalized Tajima’s D of haplotypes around human-specific fixed derived alleles. (D) Phylogeny of modern humans, Neanderthals, mice, and chickens, with their amino acid at position 200 in NOVA1 denoted [NOVA1I200 (isoleucine) and NOVA1V200 (valine)]. (E) Tertiary structure of NOVA1. Partial structure of NOVA1, showing the KH1 (yellow) and KH2 (blue) domains bound to RNA (green). The location of the variable residue at position 200 is shown in red. The structure of the KH3 domain has not been solved. RNA can simultaneously bind to both the KH1 and KH2 domains of NOVA1.
