Figure 3:
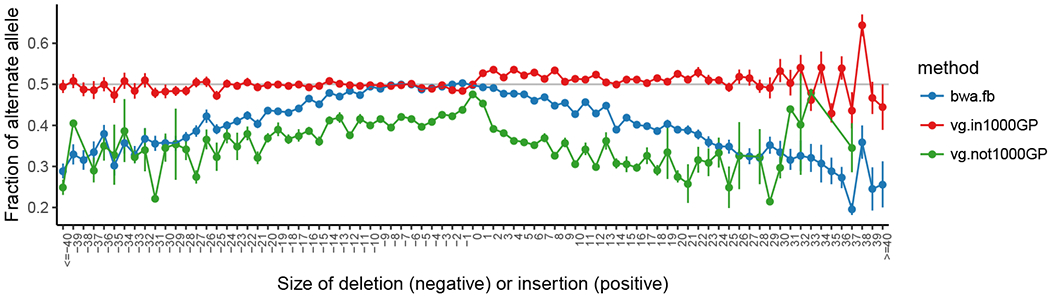
The mean alternate allele fraction at heterozygous variants in HG002/NA24385 validated in the Genome in a Bottle truth set (148) as a function of deletion or insertion size (SNPs at 0). Error bars are ± 1 s.e.m. Blue points show the allele balance metric across allele lengths for alignments with bwa mem (83) and variant calls made by freebayes (50). Variant calls were made with alignment to a variation graph built from the 1000 Genomes Project variants (1), followed by variant calling in VG. These are divided into two groups: calls at variants in the graph used for the alignment (red) and those to variants that are private to HG002 (green). Reprinted from (51).
