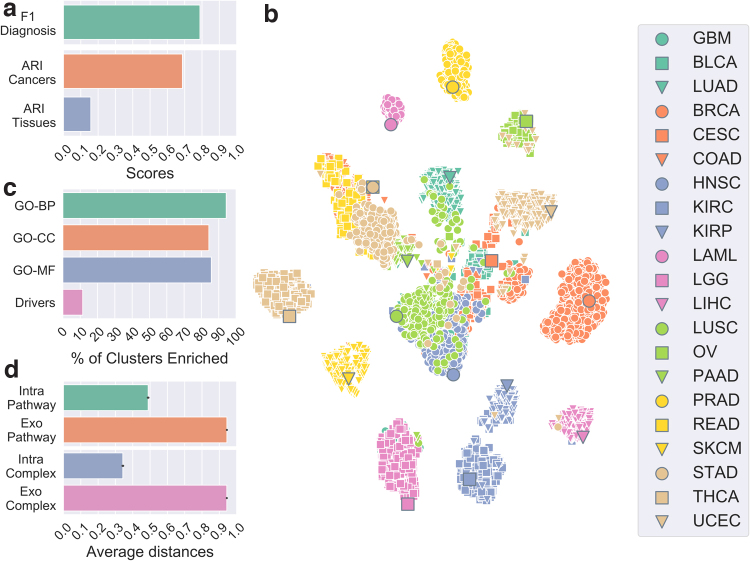
FIG. 2.
(a) Macro-F1 score quantifying the relationship between a patient and its cancer type (green) and ARI measuring the link between patient clustering for each model and cancer type labeling (orange) and tissue-sampled labeling (blue). (b) t-SNE plot representing the embedding of patients and cancer types in the latent space. The larger circled markers correspond to the embeddings of cancer types, and the smaller ones represent the embeddings of patients. Colors and shapes indicate cancer types (see Supplementary Table S1 for abbreviations meanings). (c) Percentages of gene clusters enriched in GO-BP, GO-CC, GO-MF, and driver annotations. (d) Average cosine distance between genes and associated pathways and complexes (intrapathway and intracomplex) and nonassociated pathways and complexes (exo-pathway and exo-complex). ARI, adjusted rand index; BP, biological processes; CC, cellular component; GO, Gene Ontology; MF, molecular function; t-SNE, T-distributed stochastic neighbor embedding.