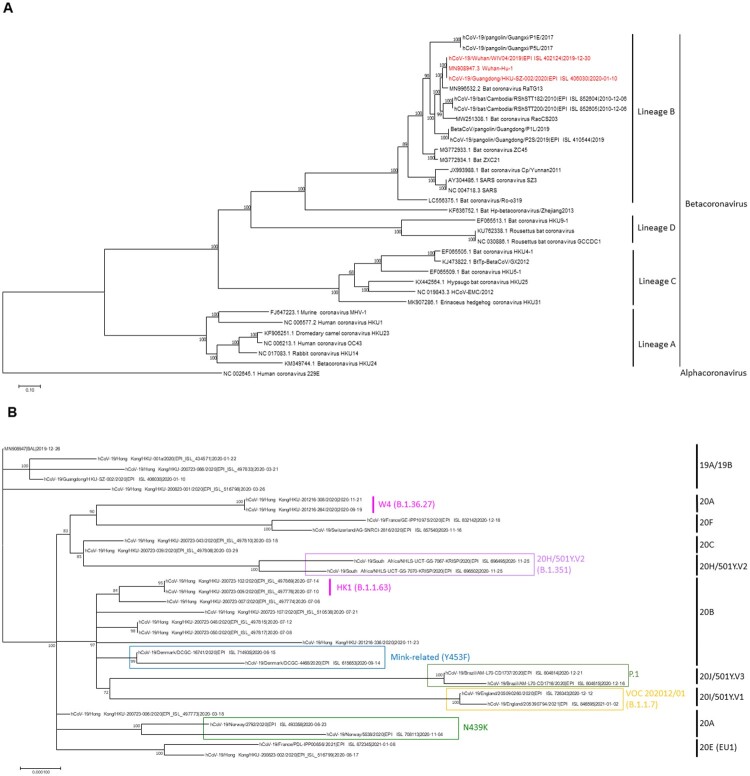
Figure 2.
(A) Whole genome phylogenetic tree of betacoronaviruses. The tree was constructed by maximum likelihood method with the best-fit substitution model GTR + F+R5 using IQTree2. Bootstrap values were calculated by 500 trees. SARS-CoV-2 are highlighted in red. Human coronavirus 229E (NC_002645) was used as outgroup. (B) Whole genome phylogenetic analysis showing different clades of SARS-CoV-2. The tree was constructed by maximum likelihood method with the best-fit substitution model TIM2+F + I using IQTree2. Bootstrap values were calculated by 500 trees. Clade information as inferred by Nextstrain or Pango lineage are shown. HK1 is the predominant lineage found during the 2020 summer peak in Hong Kong, while W4 is the predominant lineage that is found in almost all local cases in Hong Kong since November 2020. The reference genome Wuhan-Hu-1 (GenBank accession number MN908947.3) is used as the root of the tree.