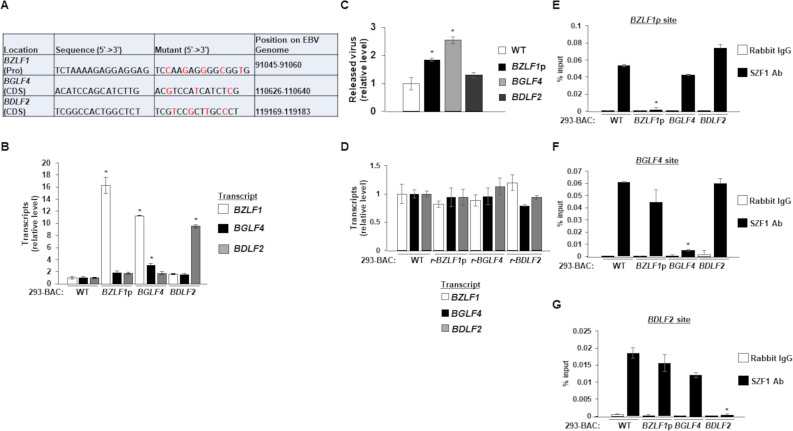
Fig 3. Mutations in candidate SZF1-binding sites derepress EBV lytic genes.
(A) Synonymous point mutations were made in candidate SZF1-binding sites on the p2089 BACmid via red recombineering. Mutant residues are shown in red. (B) After transfection and hygromycin selection (~ 2 weeks later), 293-BAC cells harboring wild-type p2089 BACmid or BACmids with mutant SZF1-binding sites were harvested for RNA extraction and RT-qPCR analysis for relative expression of EBV lytic genes (from each kinetic class: BZLF1, immediate early; BGLF4, early; BDLF2, late), compared to wild-type BAC sample. (C) Supernatants from 293-BAC cells harboring p2089-BACs bearing BZLF1 promoter, BGLF4 coding sequence, or BDLF2 coding sequence SZF1-binding site mutations were harvested and analyzed via qPCR for relative amounts of released DNase-resistant virus compared to the wild-type 293-BAC sample. (D) 293-BAC cells harboring wild-type p2089-BAC or p2089 BACmids that underwent reversion (r) mutations for their respective SZF1-binding sites were tested by RT-qPCR of lytic genes BZLF1, BGLF4, and BDLF2 relative to wild-type 293-BAC. (E-G) SZF1-ChIP was performed on wild-type 293-BAC samples or 293-BACs harboring SZF1-binding site mutations; precipitated chromatin was analyzed via qPCR using primers to amplify PCR products flanking the putative BZLF1 promoter sequence (E), the BGLF4 coding sequence site (F), or the BDLF2 coding sequence site (G). ChIP-PCR results were analyzed relative to 1% input and displayed as percent input. Data represent averages of three independent experiments; error bars, SEM; *, p ≤ 0.05.