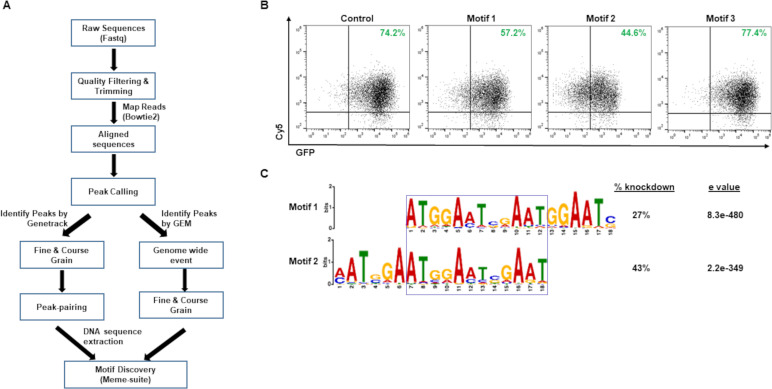
Fig 6. Evaluation of SZF1-footprint derived motifs on the B cell genome.
(A) Informatic workflow for SZF1 motif discovery. (B) Informatically-derived putative SZF1-binding motif consensus sequences were cloned into a pEGFP-N1 vector and transfected into HEK293T cells. GFP expression was assessed relative to control pEGFP-N1 vector lacking putative SZF1 motif consensus sequences; Cy5-non-targeting siRNA was co-transfected to monitor transfection efficiency. Percent GFP+ cells from three of 27 motif-consensus-sequence-bearing cassettes are displayed in green; only consensus sequences derived from motifs 1 and 2 repressed GFP expression, and motif 3 is a consensus sequence representative of the remaining 24 motifs. (C) SZF1 motifs derived from Meme-suite. Motif logos of the two motifs capable of repressing GFP in (B) were aligned and overlap demarcated by a box. Percent GFP knockdown in (B) and statistical significance of motif determined using Meme-suite are shown.