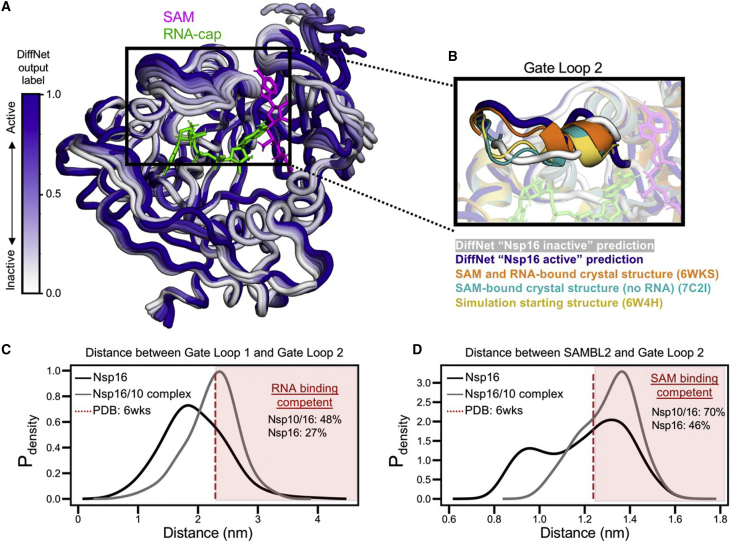
Figure 2.
Nsp10 binding shifts Nsp16’s conformational ensemble, increasing its propensity to adopt structural states that are ligand binding compatible. (A) 10 structures of Nsp16 that represent the DiffNet prediction changing from inactive to active (white to purple). The DiffNet output label scales from 0 to 1 (white to purple) reflecting the extent the DiffNet predicts a structure to be associated with Nsp16 activation. (B) Comparison of the DiffNet-predicted active and inactive states (purple plus white, respectively) to the starting simulation state (yellow), a known SAM- and RNA-bound structural state (orange), and a known SAM- (but not RNA-) bound state (teal). All structures are aligned to 6wks (orange). (C) Probability-weighted distance distribution between RNA-binding gate loops 1 and 2, comparing monomeric Nsp16 (black) to the Nsp10-Nsp16 complex (gray). (D) Probability-weighted distance distribution between SAM-binding loop 2 and gate loop 2, comparing monomeric Nsp16 (black) to the Nsp10-Nsp16 complex (gray). For (C) and (D), the distance for a SAM- and RNA-bound crystal structure is also plotted (red dotted line). To see this figure in color, go online.