Abstract
Mutations in BRCA1/BARD1’s E3 ubiquitin ligase RING domains predispose carriers to breast and ovarian cancers. We present the first structure of the BRCA1/BARD1 RING heterodimer with the E2 enzyme UbcH5c bound to its cellular target, the nucleosome, along with biochemical data that explain how the complex selectively ubiquitylates lysines 125/127/129 in the flexible C-terminal tail of H2A in a fully human system. The structure reveals that a novel BARD1-histone interface couples to a repositioning of UbcH5c compared to the structurally similar PRC1 E3 ligase Ring1b/Bmi1 that ubiquitylates H2A Lys119 in nucleosomes. This interface is sensitive to both H3 Lys79 methylation status and mutations found in cancer patients. Furthermore, NMR reveals an unexpected mode of E3-mediated substrate regulation through modulation of dynamics in the C-terminal tail of H2A. Our findings provide insight into how E3 ligases preferentially target nearby lysine residues in nucleosomes by a steric occlusion and distancing mechanism.
Introduction
Germline mutations in BRCA1 and BARD1 predispose carriers to breast and ovarian cancer. BRCA1/BARD1 is a large heterodimeric complex best characterized for its role in DNA double-stranded break (DSB) repair by homologous recombination1,2. Its only known enzymatic activity is as an E3 ubiquitin (Ub) ligase, encoded by the heterodimeric N-terminal RING domains of BRCA1 and BARD13,4. A variety of putative cellular targets for BRCA1/BARD1-dependent E3 ligase activity have been identified, but the functional significance for most is not well understood. Furthermore, a role for the E3 ligase activity of BRCA1/BARD1 in DNA damage repair and tumorigenesis remains controversial5–12.
Nucleosomal histone H2A was recently discovered as a bona fide substrate for BRCA1/BARD1-dependent E3 ligase activity13–15. The heterodimeric RING domains are sufficient for preferential mono-ubiquitylation of lysine residues 125/127/129 on the flexible extreme C-terminal tail of canonical H2A14 and Lys123 in nucleosomes containing the macroH2A1 isoform16. The non-E2 binding RING domain of BARD1 is required for this activity, and cancer-predisposing mutations in this domain specifically abrogate nucleosome binding and ubiquitylation17. BRCA1/BARD1-dependent H2A ubiquitylation contributes to DNA DSB repair by homologous recombination11,18 and acts as a mark of transcriptional repression15,17.
Two other RING E3-ligases, RNF168 and the Ring1b/Bmi1 complex, bind to the nucleosome via their RING domains and facilitate mono-ubiquitylation of distinct H2A lysine residues. RNF168 ubiquitylates H2A residues Lys13/15 in the N-terminal region of H2A19,20, a signal that recruits the DSB repair factor 53BP1 to sites of damage to facilitate repair by non-homologous end joining21,22. The heterodimeric E3 ligase Ring1b/Bmi1, a component of the Polycomb repressive complex 1 (PRC1), directs ubiquitylation of H2A Lys119 in nucleosomes23,24. This modification functions in transcriptional regulation25 and may also contribute to the regulation of DNA DSB repair26. A crystal structure of the Ring1b/Bmi127 and an NMR-based model of the RNF16828 E3-E2/nucleosome complexes reveal how these E3s direct site-specific H2A ubiquitylation. Surface-exposed polar residues in the RING domains orient the E3s on the face of the nucleosome, recognizing distinct regions of the H2A/H2B acidic patch. This directs the RING-bound E2 enzymes towards opposite ends of the nucleosome disc surface, placing the E2 active site directly over the H2A lysine residues targeted for modification. For RNF168 and Ring1b/Bmi1, the target lysines are located on the boundary of ordered regions of H2A ~70 Å away from each other across the nucleosome disc surface (Fig. 1a, b).
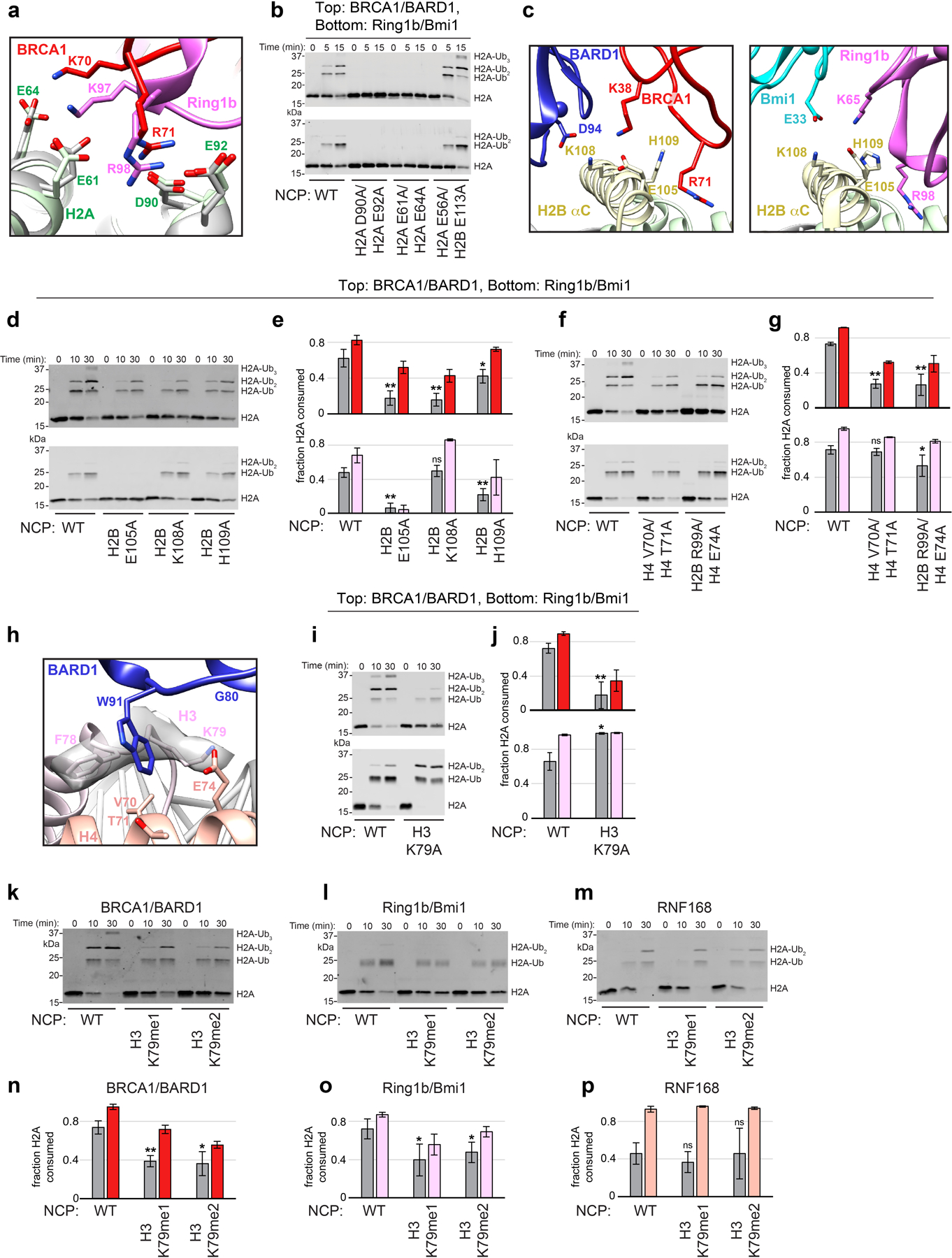
Figure 1. RING E3 ligases ubiquitylate H2A at distinct sites on the nucleosome.
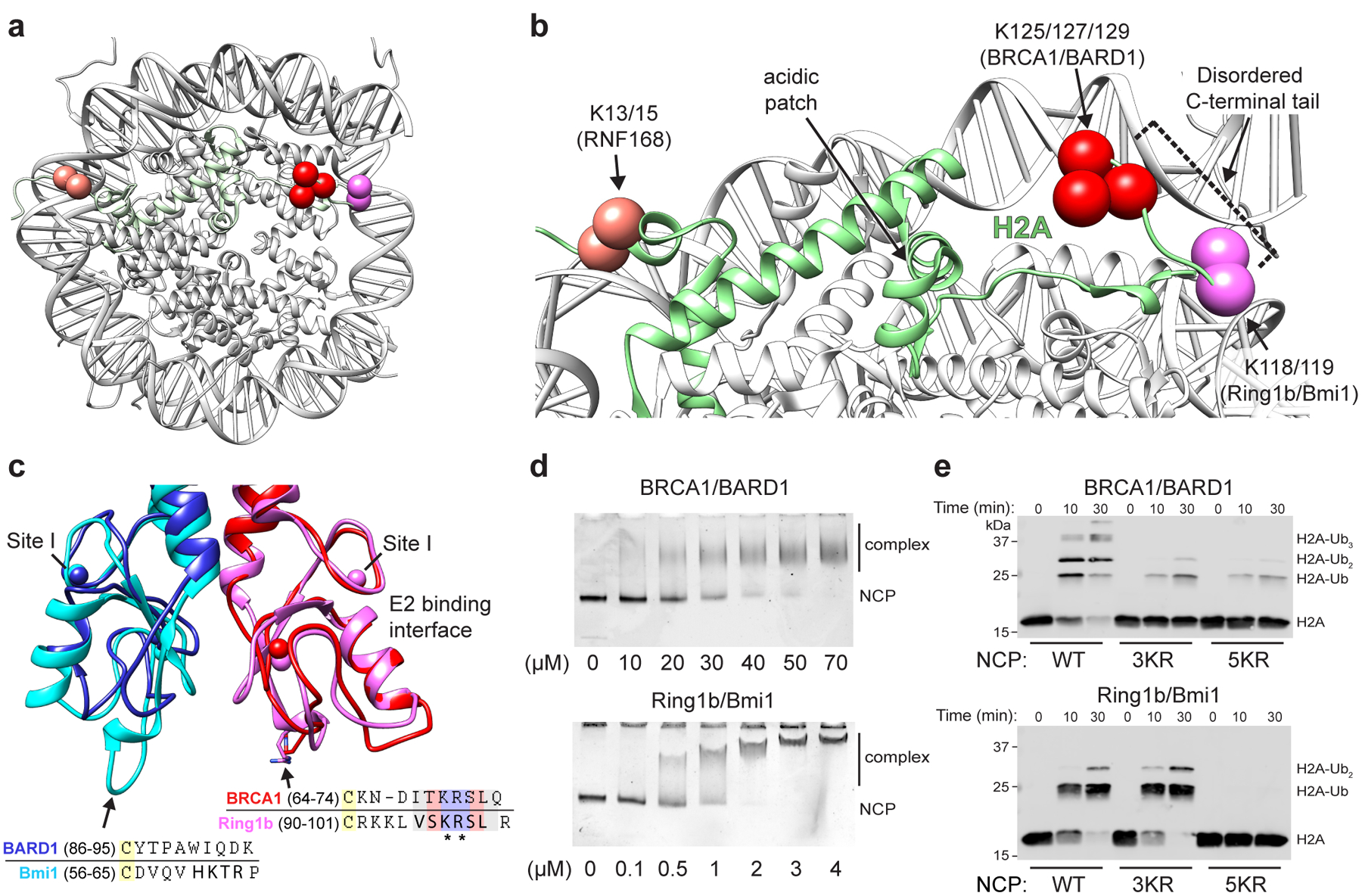
(a) Location of primary target lysine residues (Cαs, colored spheres) for BRCA1/BARD1 (K125/127/129, red) Ring1b/Bmi1 (K118/119, magenta), and RNF168 (K13/15, salmon) on one face of the nucleosome core particle (NCP; PDB: 1KX5). (b) Close-up of H2A (green) from panel a. The H2B αC helix is hidden for clarity. Residues past H2A Lys118 are disordered and generally not observed as density in X-ray or cryo-EM structures. (c) Structural and sequence alignment of BRCA1/BARD1 (PDB: 1JM7) and Ring1b/Bmi1 (PDB: 2CKL) RING domains with arrows pointing towards the nucleosome binding regions of Ring1b and Bmi1. Bolded residues in the sequence alignment are primary Ring1b and Bmi1 nucleosome binding loop residues. Asterisks denote arginine anchor motif residues. Yellow highlighted cysteine residues are nearest analogous Zn2+-coordinating residues. Zinc atoms are shown in structures as colored spheres (partially overlapping) with zinc site I labeled for each RING domain. (d) Native-gel electrophoretic mobility shift assays (EMSA) measuring BRCA1/BARD1 and Ring1b/Bmi1 RING heterodimer binding to NCPs. Concentration of RING binding partner (indicated below the gels) is different between E3s. Data are representative of n=2 (Ring1b/Bmi1) or n=3 (BRCA1/BARD1) independent binding experiments. (e) Nucleosome ubiquitylation assays (Western blot for VSV-G tag on H2A) using either BRCA1/BARD1 or Ring1b/Bmi1 RING heterodimers and wild-type (WT), H2A Lys125/127/129Arg (3KR), or H2A Lys118/119/125/127/129Arg (5KR) NCP substrates. Data in panel e are representative of n=2 independent experiments. Uncropped gel/blot images in panels d and e are available as source data.
The H2A lysines targeted by BRCA1/BARD1 (125/127/129) are nearby to those targeted by Ring1b/Bmi1 (118/119) but reside in the fully disordered extreme C-terminal tail of H2A. The Ring1b/Bmi1 and BRCA1/BARD1 RING heterodimers are topologically similar and share several functional features. BRCA1, like Ring1b, directly interacts with E2~Ub conjugates and has a conserved nucleosome-binding ‘arginine-anchor’ motif required to bind the nucleosome acidic patch27 (Fig. 1c). BARD1, like Bmi1, does not interact with the ubiquitylation machinery. These two RINGs differ primarily at the base of their RING domains where Bmi1 has an extended loop that contacts the nucleosome surface. Structural comparison calls to question how these complexes differ in binding to nucleosomes (Fig. 1d) and specifically target closely positioned H2A lysine residues (Fig. 1e). Here, we report a cryo-electron microscopy (cryo-EM) structure of the BRCA1/BARD1 RING domain heterodimer with the E2 enzyme UbcH5c bound to the nucleosome. Together with biochemical and nuclear magnetic resonance (NMR) spectroscopy data, the results reveal the basis for preferential ubiquitylation of H2A Lys 125/127/129. Our findings constitute the first structural analysis of the E3 ligase domain of BRCA1/BARD1 with any of its cellular targets and provide mechanistic insight into the process of site-specific ubiquitylation by RING-type E3 ligases.
Results
Reconstitution and cryo-EM structure of the BRCA1-UbcH5c/BARD1/nucleosome complex.
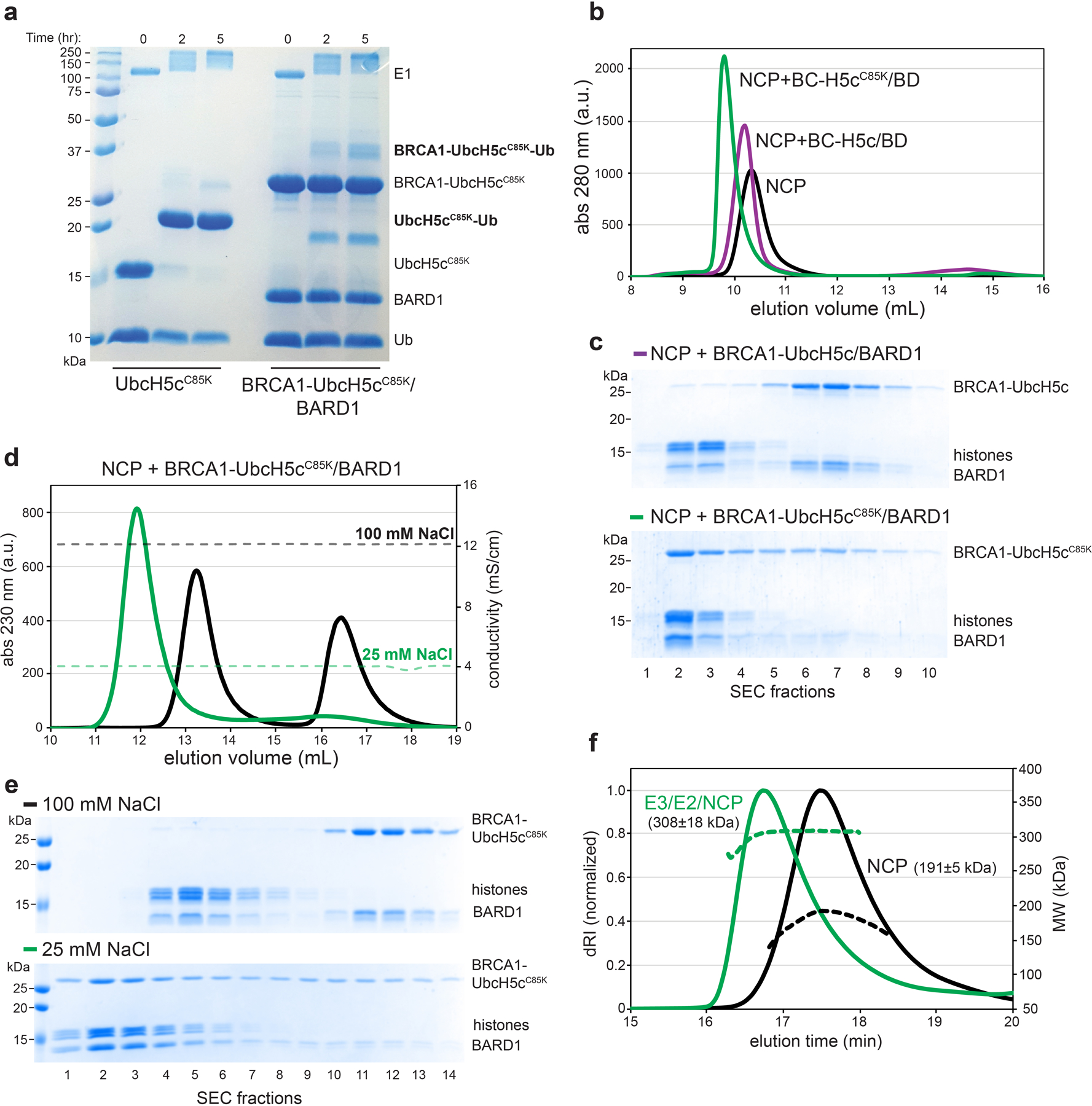
To reconstitute the BRCA1-UbcH5c/BARD1/nucleosome complex, we generated a chimeric E3-E2 enzyme where the RING domain of BRCA1 was genetically fused to UbcH5c by a flexible linker (referred to as E3-E2 module; Extended Data Fig. 1b). We took this approach to circumvent the low affinity of UbcH5c for BRCA1/BARD129 that could hinder formation of a stable ternary complex with the nucleosome. The E2 enzyme UbcH5c was chosen because it has similar activity in nucleosome ubiquitylation assays with BRCA1/BARD1 to other members of the UbcH5 (Ube2d) and Ube2e families and was used to characterize the Ring1b/Bmi127 and RNF16828 nucleosome complexes (Extended Data Fig. 1c). Importantly, the E3-E2 module retained enzymatic activity and specificity for lysine residues 125/127/129, validating its use for structural studies (Extended Data Fig. 1d, e). Consistent with a previous report27, Ub could not be added to the active site of UbcH5cCys85Lys in the context of the E3-E2 module to form a stable isopeptide-linked E3-E2-Ub moiety (Extended Data Fig. 2a). Serendipitously, we found that substitution of the E2 active site cysteine to lysine stabilized the E3-E2/nucleosome interaction, allowing for purification of the complex by size exclusion chromatography (SEC; Extended Data Fig. 2b, c). This mutation appears to increase the association of the complex through non-specific, weak electrostatic interactions as a modest increase in ionic strength is sufficient to disrupt binding (Extended Data Fig. 2d, e). The complex was observed to have an approximate molecular weight consistent with an expected 2:1 binding stoichiometry by SEC coupled to multi-angle light scattering (Extended Data Fig. 2f). Owing to the stronger observed interaction of this complex, it was used for subsequent cryo-EM and NMR studies.
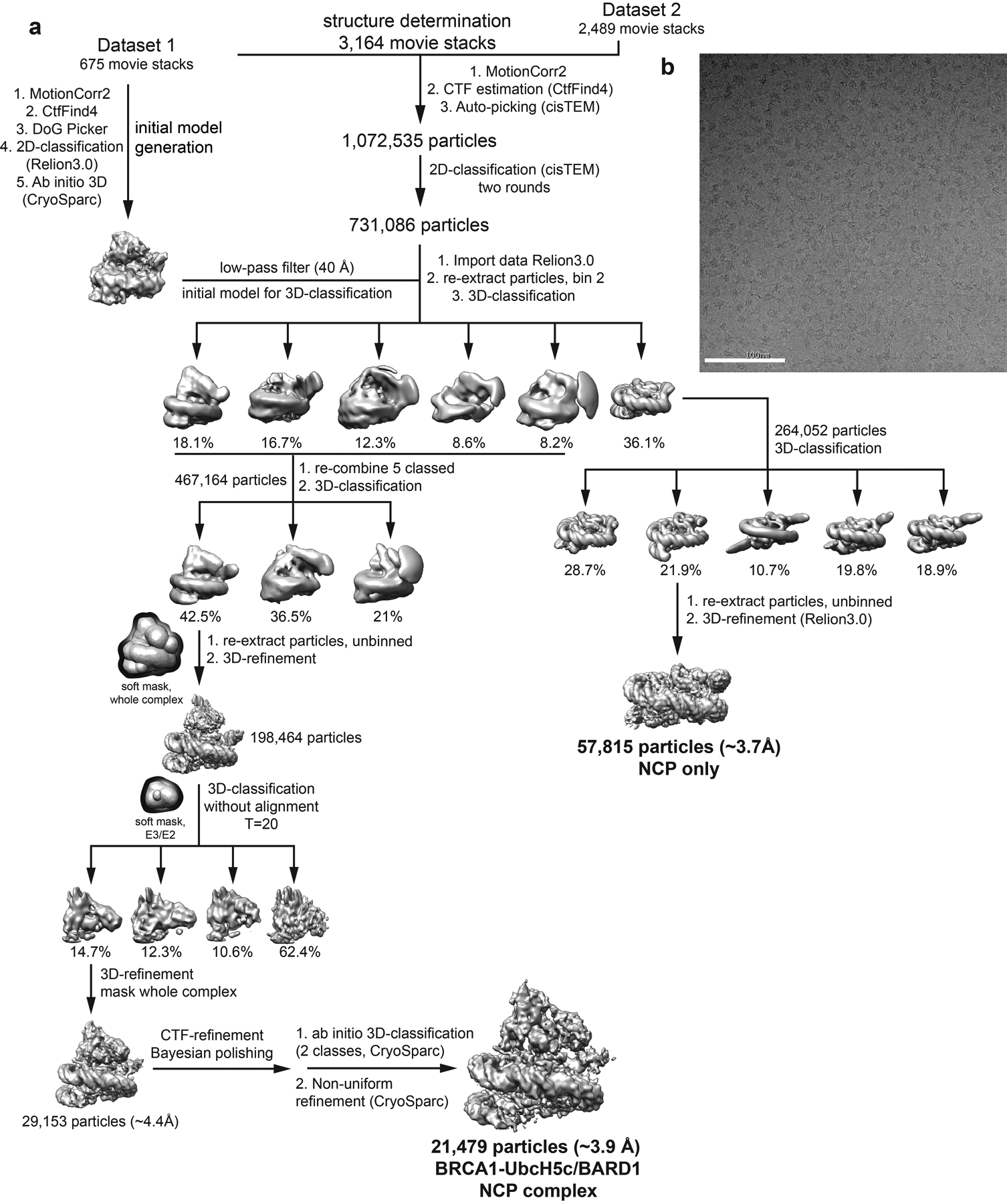
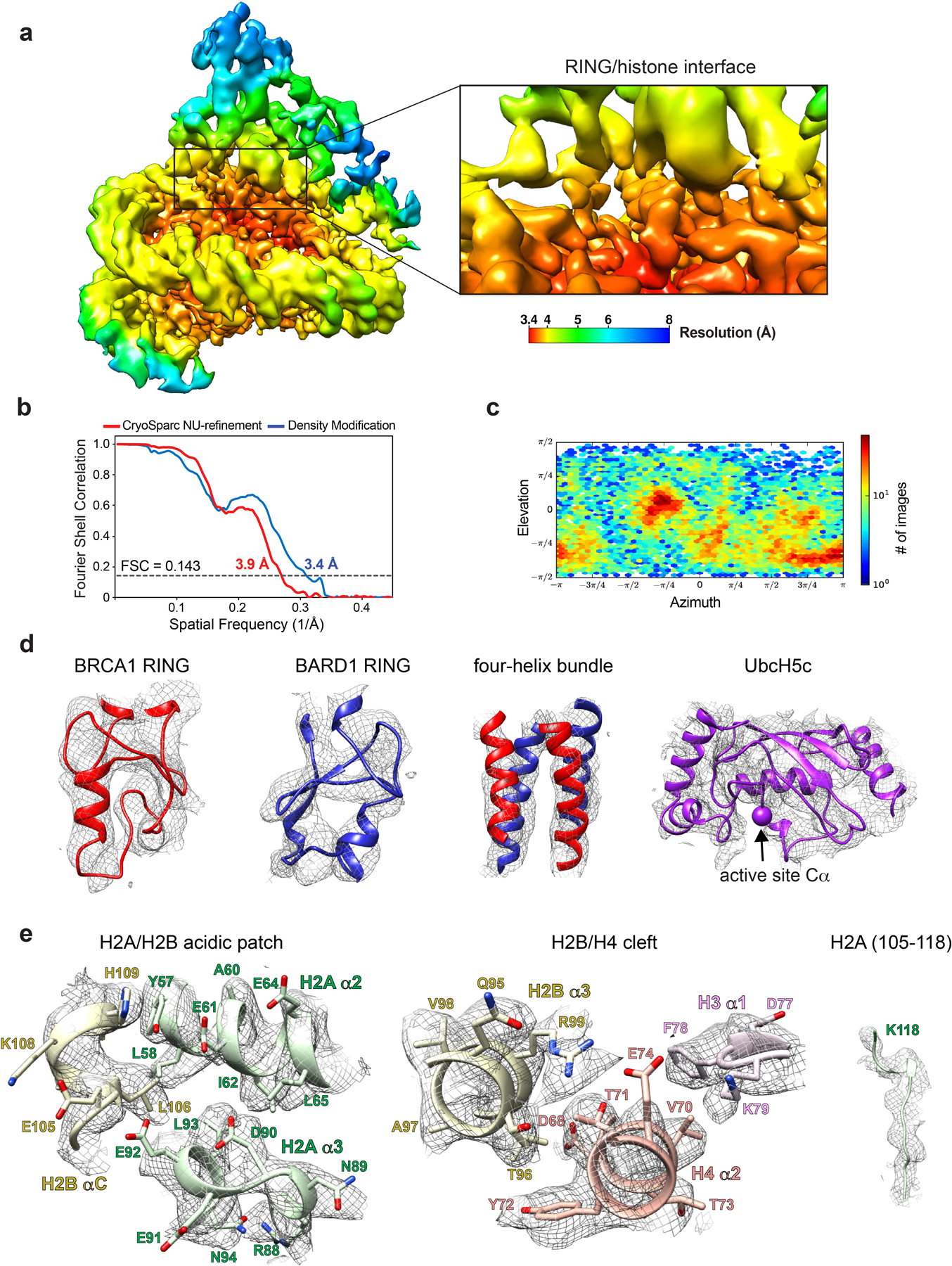
Although the complex co-purified on SEC, the E3-E2 module dissociated from the nucleosome upon cryo-EM grid preparation and was therefore stabilized by light crosslinking with glutaraldehyde immediately prior to vitrification. We obtained a cryo-EM reconstruction of the bound complex that refined to a global resolution of ~3.9 Å (3.4–7.8 Å) (Fig. 2a, Extended Data Figs. 3 and 4, Table I). The structure shows the E3-E2 module associated with one face of the nucleosome, consistent with recent cryo-EM structures of similar complexes expected to bind with 2:1 stoichiometry30,31. The polypeptide backbone is traceable in the density map for ordered regions of the nucleosome and the BRCA1/BARD1 RING domains. Density for the E2 enzyme UbcH5c is noisier with missing regions in the C-terminal end furthest from the E3-E2 interface (Fig. 2a and Extended Data Fig. 4). Importantly, map quality was sufficient for unambiguous docking with full or partial side chain density for many key interface residues. Model building was accomplished by a combination of homology modelling, de novo structure prediction, rigid-body docking, and all-atom refinement in Rosetta32,33 (Fig. 2b and Table I). The resulting structure shows that the BRCA1/BARD1 RING domains straddle the H2B αC helix, binding to both the H2A/H2B acidic patch and H2B/H4 cleft (Fig. 2b). UbcH5c is bound to the BRCA1 RING in a canonical manner and is tilted away from the histone surface, terminating near the DNA ends without making specific interactions with the nucleosome.
Figure 2. Cryo-EM structure of the BRCA1-UbcH5c/BARD1/nucleosome complex.
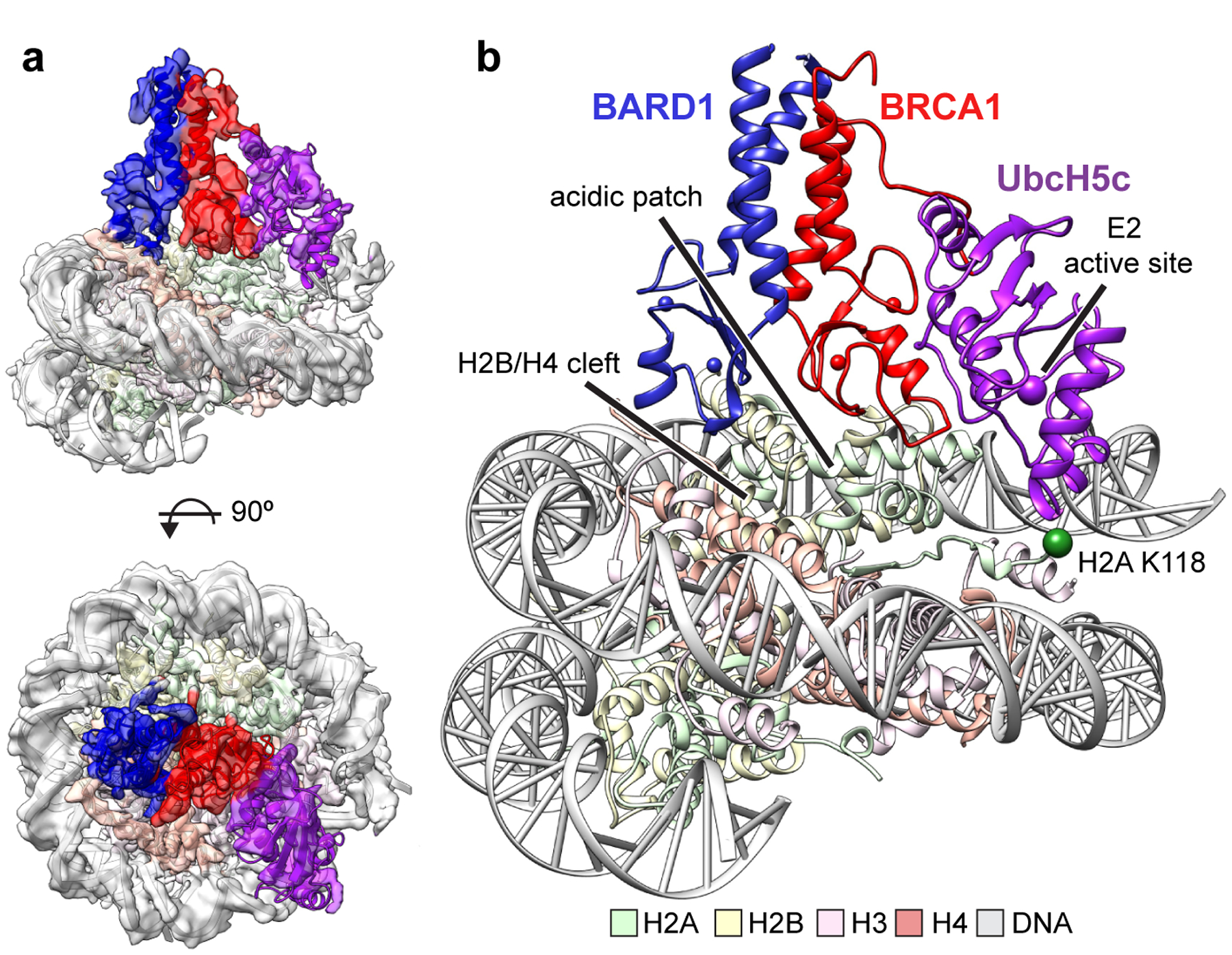
(a) Cryo-EM density of the BRCA1-UbcH5c/BARD1/NCP complex (semi-transparent surface) with fit atomic model. Density map is colored coded by chain using the same scheme as in panel b. (b) Atomic model of BRCA1-UbcH5c/BARD1/NCP complex. The Cαs of H2A Lys118 and the active site position of UbcH5c (Lys85) are depicted at spheres.
Table I.
Cryo-EM data collection, refinement and validation statistics
| BRCA1-UbcH5c/BARD1 nucleosome complex (EMD-22581, PDB 7JZV) | |
|---|---|
| Data collection and processing | |
| Magnification | 130,000 |
| Voltage (kV) | 300 |
| Electron exposure (e–/Å2) | 90 |
| Defocus range (μm) | −0.7 to −2.7 |
| Pixel size (Å) | 1.05 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 1,072,535 |
| Final particle images (no.) | 21,479 |
| Map resolution (Å) | 3.9 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 3.4–7.8 |
| Refinement | |
| Map sharpening B factor (Å2) | −85 |
| Model composition | |
| Non-hydrogen atoms | 14,339 |
| Protein residues | 1,093 |
| Nucleotides | 278 |
| Ligands | Zn2+(4) |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.28 |
| Validation | |
| MolProbity score | 0.80 |
| Clashscore | 1.02 |
| Poor rotamers (%) | 0.22 |
| Ramachandran plot | |
| Favored (%) | 98.32 |
| Allowed (%) | 1.49 |
| Disallowed (%) | 0.19 |
BRCA1/BARD1 RING-histone interactions.
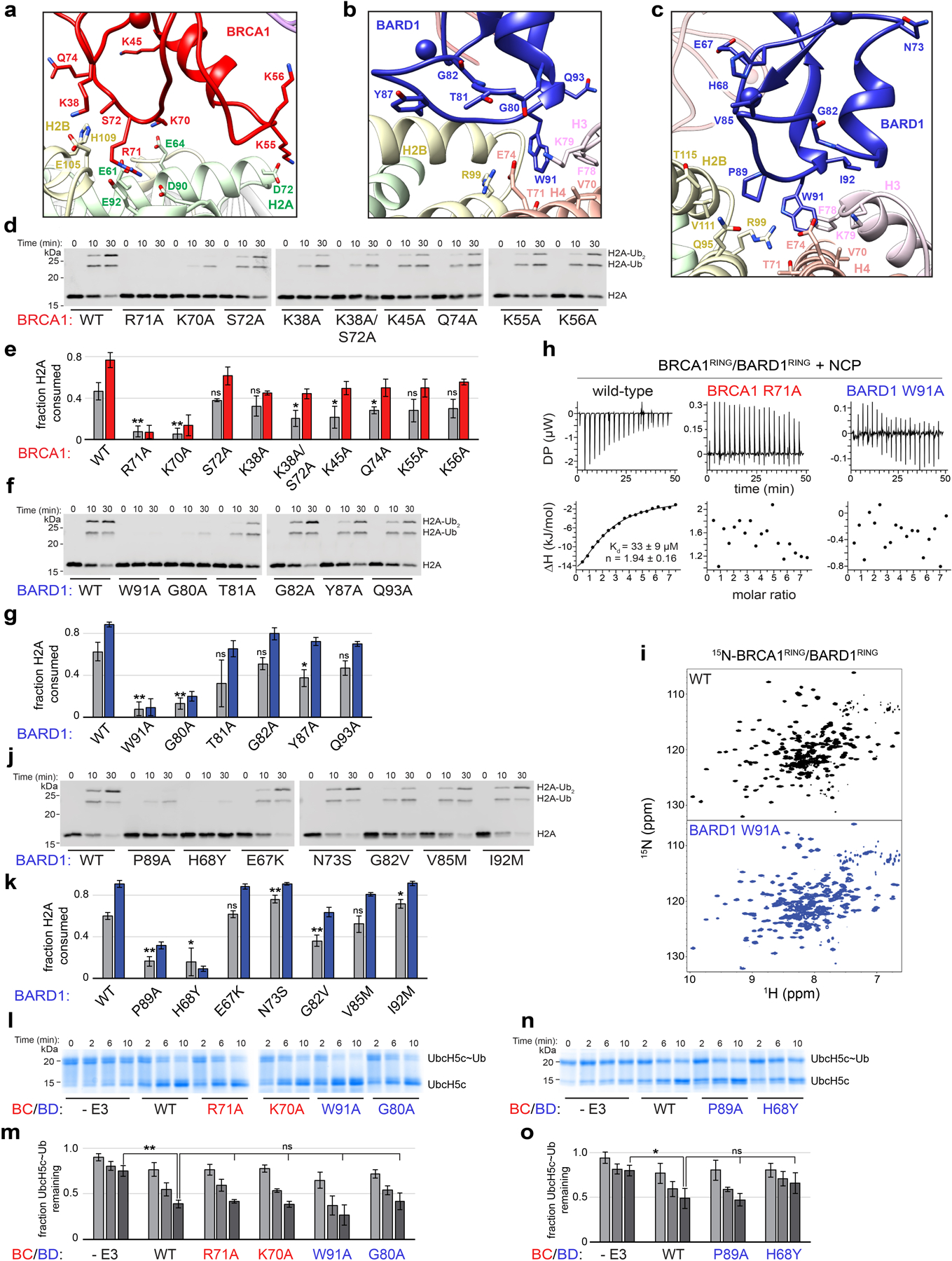
The BRCA1 RING domain shares a common nucleosome binding motif with Ring1b that is required for nucleosome ubiquitylation in in vitro activity assays27. Central to this motif is the BRCA1 Arg71 side chain that is observed to insert into a pocket in the nucleosome acidic patch composed of side chains from H2A residues Glu61, Asp90, and Glu92 (Fig. 3a). Our model predicts that the conserved neighboring Lys70 of BRCA1 interacts with the side chain of H2A Glu64 that is also observed in the Ring1b-histone interface. BRCA1 Arg71Ala was the only mutant tested that abrogated detectable nucleosome ubiquitylation and the Lys70Ala mutant was also severely affected (Fig. 3c). Binding to the nucleosome was also not detected for the BRCA1 Arg71Ala mutant using isothermal titration calorimetry (ITC; Fig. 3e, Extended Data Fig. 5h). These data identify Arg71 as the ‘arginine-anchor’ common to many chromatin factors34. Mutation of BRCA1 RING domain residues surrounding Lys70 and Arg71 had diminishing effects on decreasing nucleosome ubiquitylation (Extended Data Fig. 5a, d, e).
Figure 3. BRCA1/BARD1 RING-histone interactions.
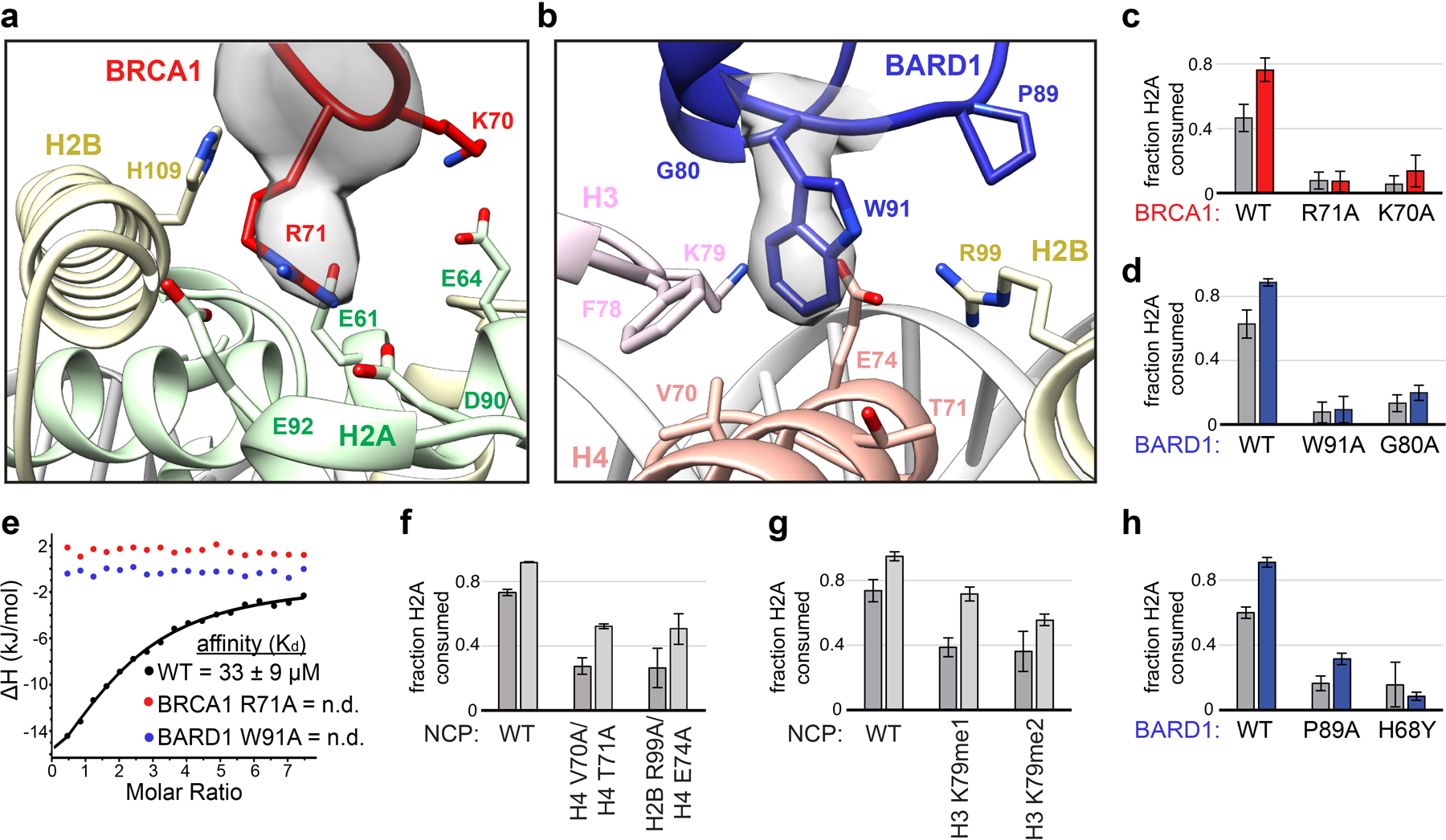
(a) Close-up view of the BRCA1-histone interface with Arg71 side chain density shown. (b) Close-up view of the BARD1-histone interface with Trp91 side chain density shown. Relevant side chains in panels a and b are shown as sticks. (c, d) Quantified nucleosome ubiquitylation assays using the indicated BRCA1/BARD1 mutants. (e) Binding of BRCA1/BARD1 (wild-type and indicated mutants) to NCPs measured by isothermal titration calorimetry (ITC). Data are representative of n=2 experiments (n.d. = not detected). (f) Quantified nucleosome ubiquitylation assays using BRCA1/BARD1 and the indicated H2B/H4 cleft mutant NCP substrates, (g) H3 Lys79 methylation mimetic NCP substrates, and (h) BARD1 COSMIC mutants. All quantified ubiquitylation activity data show the mean; error bars are ± 1-s.d. of n=3 independent experiments for 10-minute (gray bars) and 30-minute (light gray or colored bars) time points. Data for graphs in c, d, f, g, and h are available as source data.
To further validate the BRCA1/BARD1-histone interface, we tested the activity of BRCA1/BARD1 and Ring1b/Bmi1 with nucleosomes mutated at the H2A/H2B acidic patch. Consistent with previous reports27,28, the same canonical acidic patch surface is required for BRCA1/BARD1- and Ring1b/Bmi1-dependent nucleosome ubiquitylation (Extended Data Fig. 6a, b). Residues at the N-terminal end of the H2B αC helix that line the nucleosome acidic patch are poised to make analogous interactions with residues on BRCA1/BARD1 and Ring1b/Bmi1 (Extended Data Fig. 6c). H2A ubiquitylation by BRCA1/BARD1 decreased similarly for H2B Glu105Ala and Lys108Ala nucleosome mutants. In contrast, nucleosome ubiquitylation by Ring1b/Bmi1 was nearly abrogated with an H2B Glu105Ala substrate but was less affected by the Lys108Ala and His109Ala mutants (Extended Data Fig. 6d, e). These results show that BRCA1/BARD1 and Ring1b/Bmi1 have similar yet distinct interactions on the nucleosomal surface that facilitate specific H2A ubiquitylation.
Although the BRCA1 and Ring1b RINGs are structurally similar and use a common nucleosome binding motif, the non-E2 binding RING domains of BARD1 and Bmi1 are more divergent (Fig. 1c). In the Ring1b-UbcH5c/Bmi1/nucleosome complex, Bmi1 binds to the C-terminal end of the H3 α1 helix using an extended loop that is largely conserved among the six human PCGF proteins that form functional heterodimers with Ring1b35. Our structure reveals that, lacking this extended loop, BARD1 binds to the nucleosome H2B/H4 cleft using a novel interface in which the Trp91 sidechain inserts into a pocket lined by residues H3 Phe78/Lys79, H4 Val70/Thr71/Glu74, and H2B Arg99 (Fig. 3b). This interaction is facilitated by a conformational change that extends the loop containing Trp91 towards the histone surface (Extended Data Fig. 7a). Mutation of BARD1 Trp91 to Ala sharply reduced nucleosome ubiquitylation and binding (Fig. 3d, e and Extended Data Fig. 5h). Importantly, Trp91 is not required for the structural integrity of the BARD1 RING, as the 1H15N-TROSY-HSQC NMR spectrum of BARD1 Trp91Ala RING heterodimer resembles that of wild-type BRCA1/BARD1 (Extended Data Fig. 5i). Nucleosomes with mutations in the BARD1 Trp91 binding-pocket were significantly impaired toward BRCA1/BARD1 ubiquitylation (Fig. 3f) but had no or lesser effects with Ring1b/Bmi1 (Extended Data Fig. 6f, g). Our structure shows that H3 Lys79 lines the BARD1 Trp91 binding pocket and is positioned near the C-terminal cap of BARD1 helix 2 (Gly80/Thr81) (Fig. 3b and Extended Data Fig. 6h). Consistent with this, BARD1 Gly80Ala or H3 Lys79Ala mutants decreased nucleosome ubiquitylation activity (Fig. 3d and Extended Data Fig. 6i, j). Mutation of surrounding residues in the BARD1 RING did not confer these effects (Extended Data Fig. 5b, f, g). Unexpectedly, mutation of H3 Lys79Ala enhanced Ring1b/Bmi1-dependent nucleosome ubiquitylation, indicating unique requirements for this residue between the two E3s (Extended Data Fig. 6i, j).
As H3 Lys79 methylation is generally associated with actively transcribed chromatin36,37, we tested the activity of BRCA1/BARD1, Ring1b/Bmi1, and RNF168 for H3 Lys79me1 and Lys79me2 methyl-lysine analog nucleosomes. Ubiquitylation activity for these methylated substrates was significantly reduced for both BRCA1/BARD1 and Ring1b/Bmi1 compared to unmethylated nucleosomes, but unaffected for RNF168 that does not bind near H3 Lys79 (Fig. 3g and Extended Data Fig. 6k–p). This potential mode of histone-modification crosstalk suggests that BRCA1/BARD1-dependent nucleosome ubiquitylation may occur more frequently at non-transcribed genes. Histone H3 Lys79 methylation has also been implicated in recruiting 53BP1 for DNA DSB repair by non-homologous end joining38,39. As BRCA1/BARD1 occupancy at DSBs antagonizes 53BP1, we speculate that H3 Lys79 methylation status may further delineate chromatin substrates for these opposing DNA repair factors.
Next, we tested the nucleosome ubiquitylation activity of a series of BARD1 RING mutants found in the expert-curated Catalogue of Somatic Mutations in Cancer (COSMIC) database40. Missense mutations at BARD1 Pro89 have been reported in five patient samples. BARD1 Pro89 is in close proximity to the Trp91 interface and forms an adjacent interface with residues on the H2B α3 and αC helices (Extended Data Fig. 5c). The BARD1 Pro89Ala mutant significantly decreased nucleosome ubiquitylation activity (Fig. 3h). A patient-derived mutant of the zinc coordinating histidine residue His68Tyr also decreased nucleosome ubiquitylation. Other reported mutants from patients did not confer deleterious effects in vitro (Extended Data Fig. 5c, j, k). All BRCA1 and BARD1 mutants found to be highly defective in nucleosome ubiquitylation assays still retained intrinsic E3 ligase activity in E2~Ub lysine discharge assays, indicating that these residues play a critical role in the assembly of a productive RING/nucleosome complex (Extended Data Fig. 5l–o). Together, these data reveal the critical contributions of the non-E2 binding BARD1 RING domain in directing specific nucleosome ubiquitylation that is modulated by the methylation status of H3 Lys79 in vitro, and also uncovers potential disease mechanisms for BARD1 RING mutations found in cancer patients.
BRCA1/BARD1 RING orientation keeps the UbcH5c active site away from H2A Lys119.
The Ring1b/Bmi1 and RNF168 RING complexes direct specific nucleosome ubiquitylation by binding to distinct histone surfaces and orienting the UbcH5c active site directly over H2A lysine residues 118/119 and 13/15, respectively. These lysine targets are on the boundary of ordered regions of H2A in the nucleosome. Because the H2A lysine residues (125/127/129) that are modified by BRCA1/BARD1 are in a fully disordered region of the C-terminal tail that is not visible by x-ray crystallography or cryo-EM, the mechanism of site-specific ubiquitylation by this E3-E2 pair is not revealed directly.
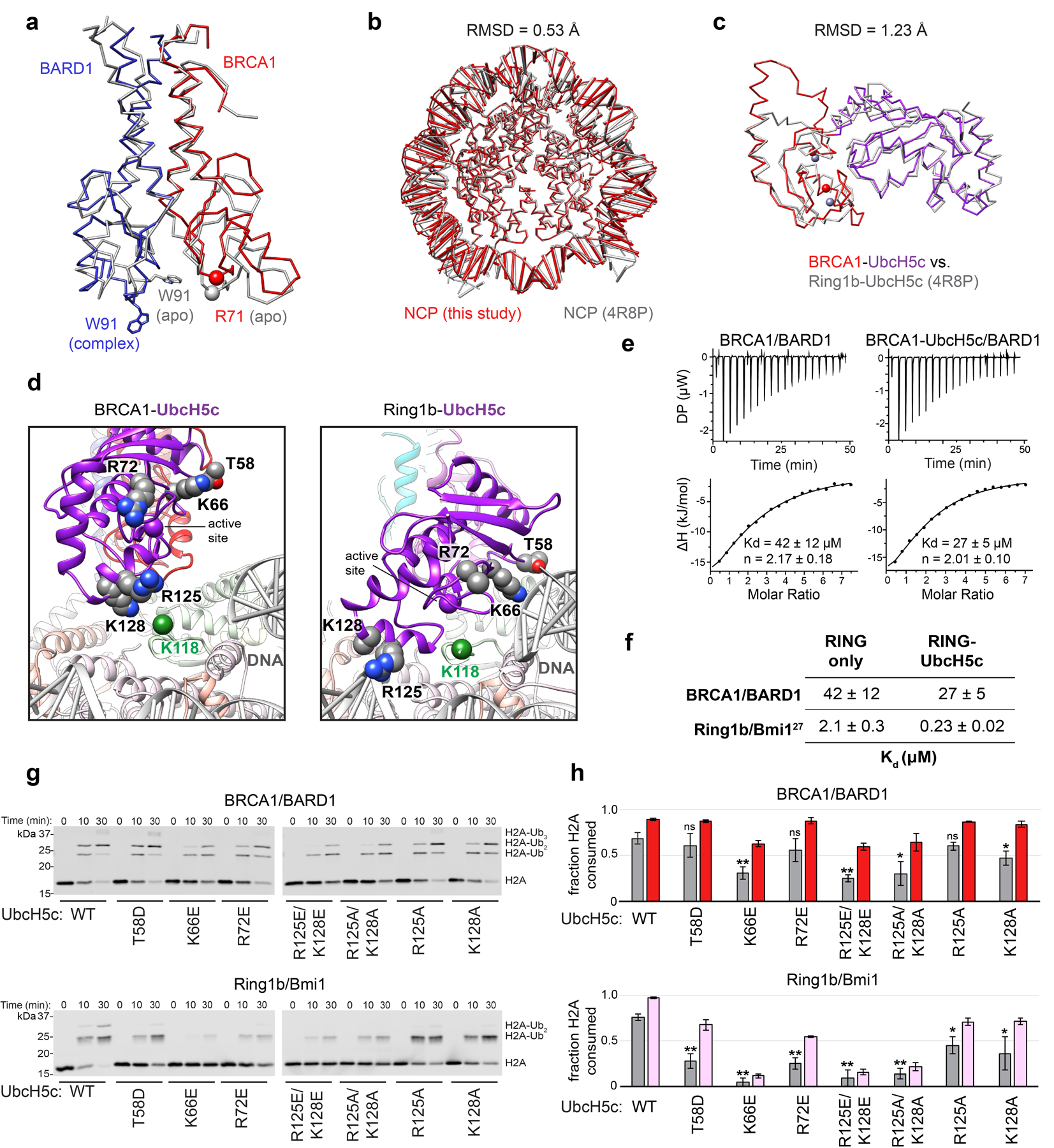
In our structure, BRCA1-bound UbcH5c is removed from the histone surface compared to its location in the Ring1b-UbcH5c/Bmi1/nucleosome complex (Fig. 4a). Although the BRCA1 and Ring1b RINGs bind similarly, the BARD1 subunit is constrained closer to the nucleosome surface and shifted ~10 Å on an axis parallel to the H4 α2 helix compared to Bmi1 (Fig. 4b). This arrangement effectively tilts the BRCA1 E2 interaction site, reorienting UbcH5c away from the nucleosome surface (Fig. 4c, d and Extended Data Fig. 7b, c). The result is that Cα-Cα distance between the E2 active site of BRCA1-bound UbcH5c and H2A Lys118 (last ordered residue in H2A C-terminus) is increased to ~19 Å from ~9Å for Ring1b-bound UbcH5c (Fig. 4a). Although elevated from the nucleosome surface, BRCA1-bound UbcH5c extends towards the DNA ends where the flexible region of the C-terminal tail of H2A emanates from the ordered histone surface. We propose that this increased distance from H2A Lys118 accounts for the observation that BRCA1/BARD1 is unable to efficiently ubiquitylate H2A Lys118/119 in the absence of its preferred lysine targets.
Figure 4. Comparison to the Ring1b-UbcH5c/Bmi1/nucleosome complex.
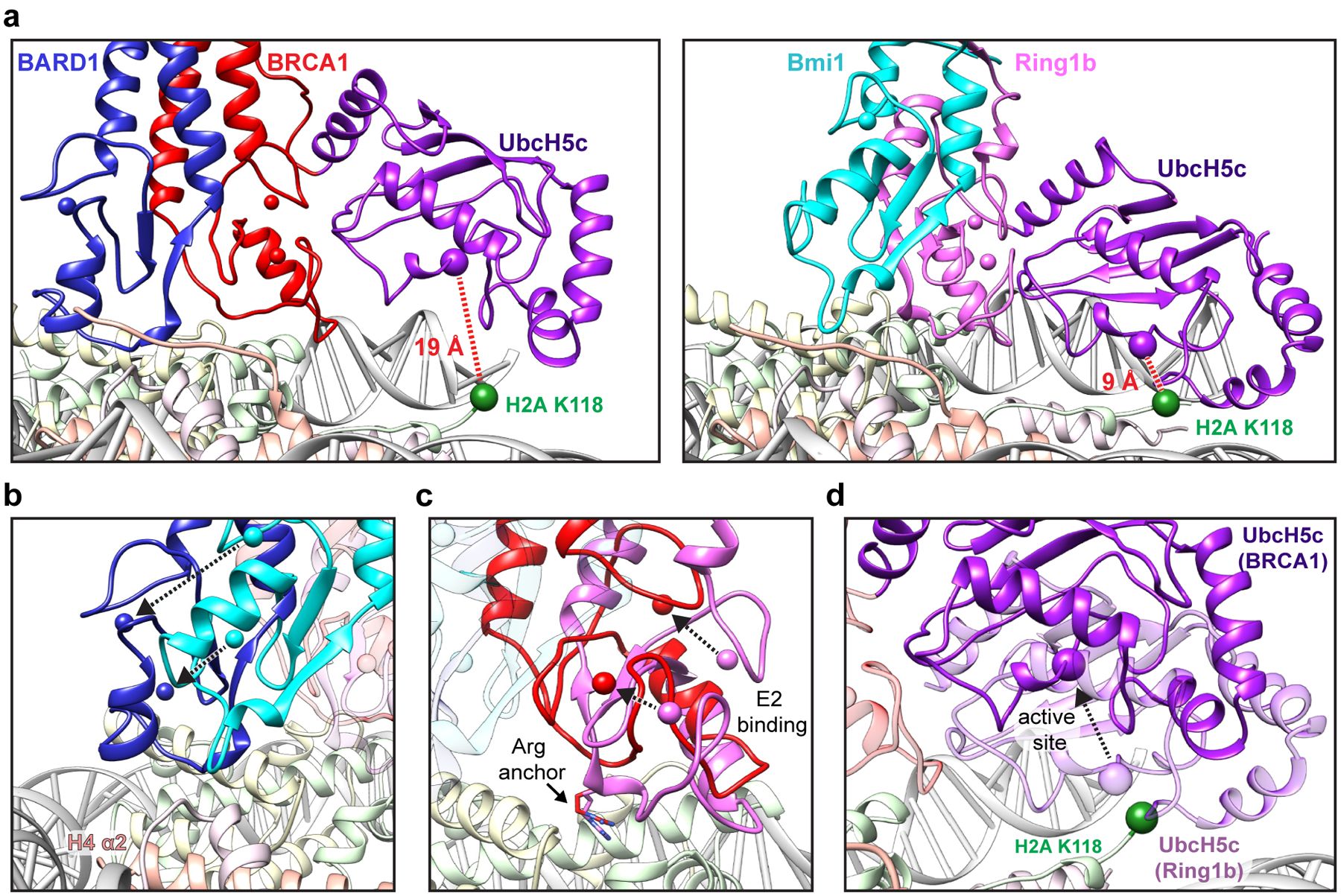
(a) Close-up views of the BRCA1-UbcH5c/BARD1/NCP complex (left panel) and Ring1b-UbcH5c/Bmi1/NCP complex (PDB: 4R8P, right panel) aligned by H2B on the bound face of the nucleosome. Complexes are shown side-by-side instead of overlaid for clarity. Distance measurements are shown between the Cα of H2A Lys118 (green sphere) and the active site position Cα Lys/Cys85 (purple sphere). All comparisons in this study are performed to the proximally bound side of the Ring1b-UbcH5c/Bmi1/NCP complex27. (b) View of the BARD1 and Bmi1 RING domains in the NCP complexes aligned by H2B. Only histones from the BRCA1-UbcH5c/BARD1/NCP model are depicted, but histone components from both complexes show excellent agreement (Extended Data Fig. 7b). The positions of analogous Zn2+ ions (colored spheres) are connected by arrows to highlight the differences in the RING domains on the histone surface. (c) Same alignment as in panel b comparing the location of the BRCA1 and Ring1b RING domains with the arginine anchor residue side chains shown. (d) Same alignment as in panel b comparing the position of BRCA1-bound UbcH5c (dark purple) to Ring1b-bound UbcH5c (light purple). The active site Cα position for each E2 is shown as a purple sphere, and the arrow between them highlights the shift of the active site of BRCA1-bound UbcH5c away from H2A Lys118 (green sphere). The Ring1b RING domain is hidden for clarity, and the BRCA1 RING is shown as partially transparent red ribbon.
Unlike Ring1b-bound UbcH5c, our structure does not predict extensive interactions between BRCA1-bound UbcH5c and nucleosomal DNA (Extended Data Fig. 7d). Consistent with this, the presence of the E2 does not substantially alter the binding affinity of the BRCA1/BARD1 RING-nucleosome interaction measured by ITC (42 ± 12 μM versus 27 ± 5 μM in the absence and presence of UbcH5c, respectively; Extended Data Fig. 7e, f). In contrast, incorporation of UbcH5c in the Ring1b/Bmi1 nucleosome complex increased the affinity ten-fold from ~2.1 μM for the RING complex alone to ~0.23 μM for the E3-E2 module27. Although the BRCA1/BARD1 and Ring1b/Bmi1 affinity measurements were conducted using different methods, the roughly twenty-fold difference in affinity between RING/nucleosome complexes is supported by EMSA binding data (Fig. 1d). To further test the observed difference in E2 placement, we made a series of mutations in surface exposed basic residues on UbcH5c that were shown to be important for nucleosome binding and ubiquitylation by Ring1b/Bmi1 and retain intrinsic amino-lysis activity27. These UbcH5c mutations impact Ring1b/Bmi1-dependent nucleosome ubiquitylation but have no or minimal effects on BRCA1/BARD1-dependent activity (Extended Data Fig. 7g, h). Maintenance of near wild-type like activity in BRCA1/BARD1-containing nucleosome ubiquitylation assays supports our model in which UbcH5c is tilted away from the nucleosome surface. This changes the position of the BRCA1-bound UbcH5c active site compared to Ring1b-bound UbcH5c active site, preventing modification of H2A Lys119, while allowing ubiquitylation of lysine residues 125/127/129 in the unstructured H2A C-terminal tail.
Lysine location in the flexible C-terminal tail of H2A dictates ubiquitylation efficiency.
The H2A lysine targets of BRCA1/BARD1 are in a flexible region of the C-terminal tail that is not observed by density in our structure. This region is less conserved between isoforms than ordered regions of the histone41 (Fig. 5a). We hypothesized that H2A residues Lys125/127/129 are optimally located on the flexible C-terminal tail of H2A to sample the UbcH5c active site at high frequency. To test this, we measured the ubiquitylation efficiency of BRCA1/BARD1 for nucleosome substrates with single lysine residues placed throughout the C-terminal tail of H2A and a substrate extended by ten residues via a flexible linker (referred to as +10-K). A sharp increase in nucleosome ubiquitylation efficiency was observed between lysines placed at positions 121 and 123 on the H2A tail (Fig. 5b, c). Ubiquitylation efficiency peaked with a single lysine at positions 125 and 127, then decreased for each subsequent position. The lysine residue at the end of a ten-residue extension was ubiquitylated with similar efficiency as a lysine at position 121. These data are in agreement with the ~19 Å linear distance measured between Cα atoms of H2A Lys118 and the active site residue of UbcH5c (Fig. 5c, d). Together, these results indicate that a lysine residue must have an optimal reach from the ordered histone surface to sample the UbcH5c active site to be efficiently ubiquitylated. Although the native lysine at position 125 is ubiquitylated with highest efficiency in vitro, only lysine residues 127 and 129 were observed to be ubiquitylated in a previously reported cellular assay14. This discrepancy may reflect a lack of detection by mass-spectrometry or additional modes of substrate regulation that are not preserved in our unmodified recombinant nucleosome substrates.
Figure 5. Lysine position along the flexible C-terminal tail of H2A dictates ubiquitylation efficiency.
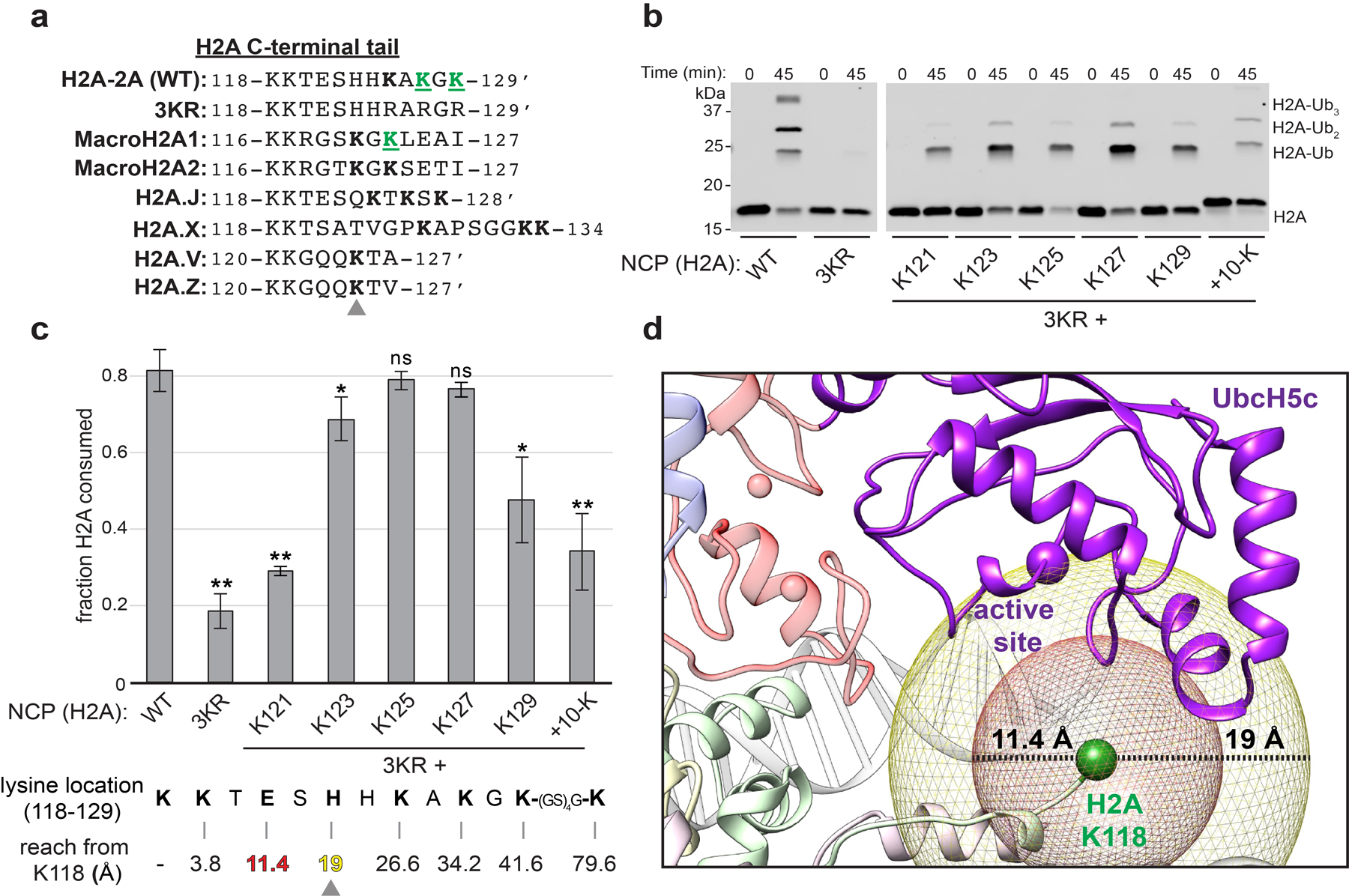
(a) Structure-based sequence alignment of H2A C-terminal tail regions from major histone H2A variants. An apostrophe denotes the C-terminus of the isoform. MacroH2A and H2Ax have extensions with additional lysine residues. Lysine residues located at positions equivalent or C-terminal to His123 in H2A-2A (WT) are bolded, with an arrow indicating this position. Green underlined residues have been identified as targets of BRCA1/BARD1 in cells by mass-spectrometry14,16. (b) Representative nucleosome ubiquitylation assay using BRCA1/BARD1 and NCP substrates containing wild-type or mutant single-lysine H2A substrates. (c) Quantified nucleosome ubiquitylation activity assays from panel b. The sequence of the H2A C-terminal tail (118–129) is below the graph, with the approximate distance from the Cα of H2A Lys118 to each single lysine position assuming an inter-residue distance equivalent to a reported contour length of 3.8 Å per amino acid42. Plotted data show the mean and error bars are ± 1-s.d. of n=3 independent experiments. P-values were calculated using a two-tailed Student’s t-test compared to wild-type (* p≤0.05, ** p≤0.005, ns = not significant). (d) The position of H2A Lys118 (Cα, green sphere) relative to the active site position UbcH5c (Cα, purple sphere). Mesh spheres are centered on the Cα of H2A Lys118 with radii corresponding to the approximate linear reach of a fully disordered polypeptide to position 121 (red) or 123 (yellow) of H2A. Uncropped gels in panel b and data for graph in c are available as source data.
The C-terminal tail of H2A is uniquely flexible in complex with BRCA1-UbcH5c/BARD1.
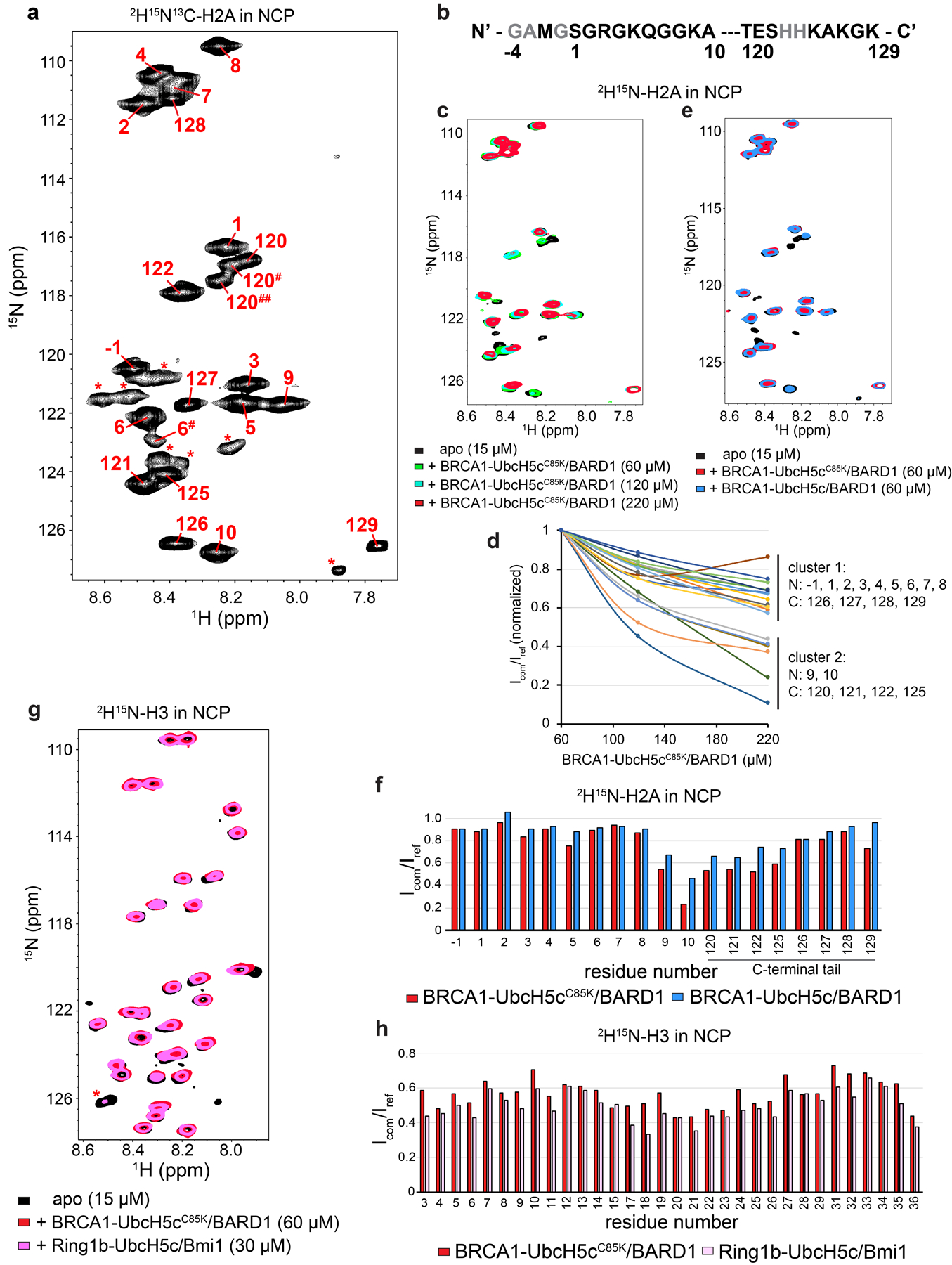
The altered position of UbcH5c in the BRCA1/BARD1 and Ring1b/Bmi1 nucleosome complexes suggests that the E2 may differentially impact interactions with the flexible C-terminal tail of H2A. Although the inherent flexibility of the H2A tails prevents observation in x-ray and cryo-EM structures, this property allows for detection by conventional NMR experiments that report on the chemical environment of backbone amide groups. We obtained backbone resonance assignments for human H2A residues 1–10, 120–121, and 125–129 in nucleosomes reconstituted with 2H15N13C-labeled H2A (Extended Data Fig. 8a, b). 1H15N-TROSY-HSQC spectra were collected for BRCA1-UbcH5cCys85Lys/BARD1 binding to 2H15N-H2A-labeled nucleosomes (Fig. 6a, left panel). Significant changes in chemical shifts were not observed. Instead, predominant spectral changes correspond to specific resonance broadening relative to unbound nucleosomes (Icom/Iref), primarily for H2A residues 120–122 and 125 (Fig. 6b). Resonances corresponding to residues 126–129 were relatively unaffected, with signal broadening similar to that measured for residues 1–10 in N-terminal tail of H2A (trendline on graph represents the mean Icom/Iref for residues −1–10). Further titration of BRCA1-UbcH5cCys85Lys/BARD1 caused uniform broadening of all observable NMR resonances indicative of non-specific binding and formation of very high molecular weight complexes (Extended Data Fig. 8c, d). Spectra collected in the presence of BRCA1-UbcH5c/BARD1 with the wild-type E2 active site revealed similar trends with lesser resonance broadening consistent with sub-stoichiometric binding or a shorter complex lifetime (Extended Data Fig. 8e, f).
Figure 6. H2A C-terminal tail dynamics in E3-E2/nucleosome complexes.
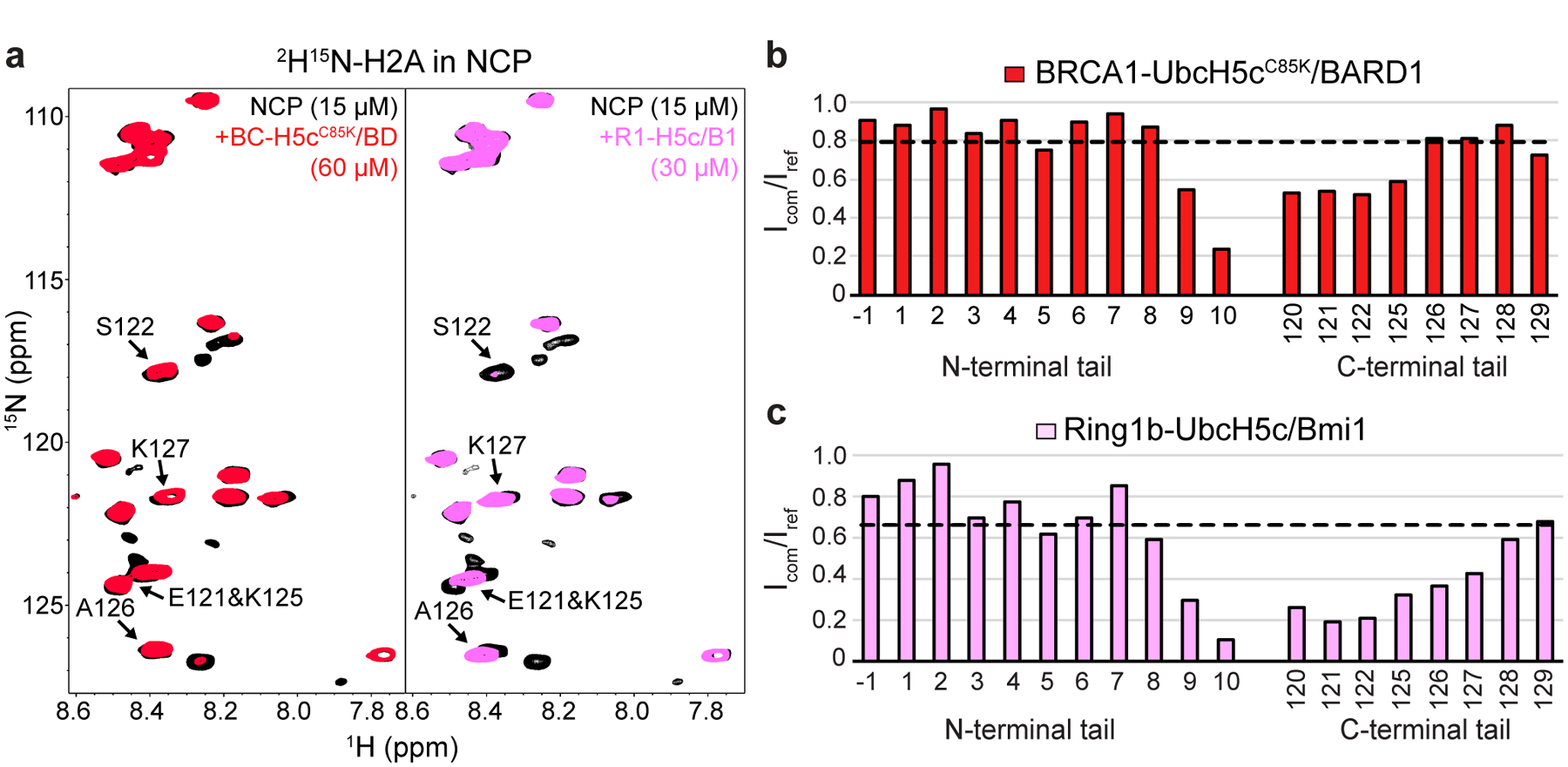
(a) Overlay of 1H15N-TROSY-HSQC spectra of 2H15N-H2A in the NCP (black, both panels), with BRCA1-UbcH5cC85K/BARD1 added (red, left panel), or Ring1b-UbcH5c/Bmi1 added (pink, right panel). Binding experiments were performed using 15 μM 2H15N-H2A NCPs with 60 μM BRCA1-UbcH5cC85K/BARD1 or 30 μM Ring1b-UbcH5c/Bmi1 added. Arrows in both panels identify signals from the H2A C-terminal tail that change position or have increased signal loss in the Ring1b-UbcH5c/Bmi1/nucleosome complex spectrum. (b) Quantification of H2A signal intensities from the BRCA1-UbcH5cC85K/BARD1/nucleosome complex in panel a comparing the bound complex spectrum to the apo reference spectrum (Icom/Iref). The trendline represents the mean broadening (Icom/Iref) from N-terminal tail residues −1–10. (c) Same analysis as in panel b for the Ring1b-UbcH5c/Bmi1/nucleosome complex. Data in panels b and c are representative of n=2 independent experiments. Data for graphs in b and c are available as source data.
Analogous spectra were collected for Ring1b-UbcH5c/Bmi1 binding to nucleosomes (Fig. 6a, right panel). Though the concentration is lower (30 μM Ring1b-UbcH5c/Bmi1 vs. 60 μM BRCA1-UbcH5cCys85Lys/BARD1) substantially greater resonance broadening was observed affecting more residues, particularly residues 120–127 (Fig. 6c). In addition, changes in chemical shifts are readily apparent for residues 121, 125, and 126, suggesting that Ring1b-UbcH5c/Bmi1 binding alters the environment of these residues to a greater extent in the nucleosome complex. Some of these effects may be attributed to tighter binding of the Ring1b-UbcH5c/Bmi1 complex. However, complementary experiments with nucleosomes reconstituted with 2H15N-labeled H3 showed robust and similar loss in intensity for both complexes due to global resonance broadening at these concentrations (Extended Data Fig. 8g, h). As the two E3-E2 complexes have approximately the same molecular weight, general signal broadening due to increases in complex molecular size was expected to be similar. These data are consistent with current models where UbcH5c in the Ring1b/Bmi1/nucleosome complex, located closer to the nucleosome surface and the emerging H2A tail, would have a greater impact on tail flexibility. Formation of the BRCA1-UbcH5c/BARD1/nucleosome complex does somewhat restrict the flexibility of the H2A C-terminal tail closest to Lys119 on the ordered histone surface, while the extreme C-terminal region where lysine targets 127/129 are located retains native-like flexibility.
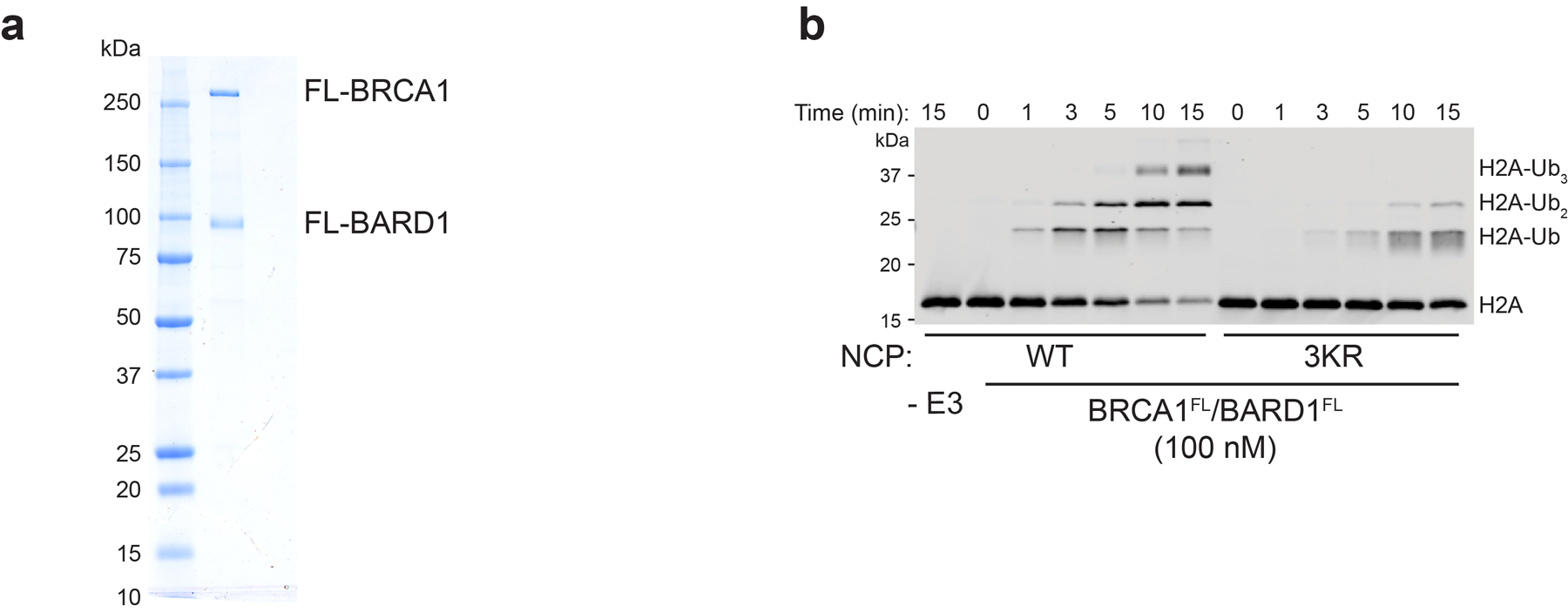
Full-length BRCA1/BARD1 has increased activity and affinity for nucleosomes
Although the RING domains of BRCA1/BARD1 are sufficient for binding and specific nucleosome ubiquitylation, the modest affinity of the BRCA1/BARD1 RING heterodimer for unmodified nucleosome substrates suggests that additional interactions may play a role. It has recently been suggested that the C-terminal region of BARD1 contributes to chromatin binding to facilitate DNA DSB repair43,44. Furthermore, both BRCA1 and BARD1 harbor DNA binding regions and interact with a variety of other chromatin-related factors45,46 (Fig. 7a). Using highly purified full-length human BRCA1/BARD1 that shows specificity for H2A Lys125/127/129 in an unmodified nucleosome substrate, we observe faster H2A ubiquitylation kinetics and a greater than 100-fold increase in nucleosome binding affinity compared to the RING heterodimer (Fig. 7b, c and Extended Data Fig. 9). This observation confirms that regions outside the RING domains of BRCA1/BARD1 contribute to interactions with nucleosomes in the context of H2A ubiquitylation. These auxiliary interactions are likely to provide additional layers of regulation and recruitment in vivo. Furthermore, it suggests that cancer-predisposing mutations outside of the RING domains may indirectly affect nucleosome-specific E3 ligase functions of BRCA1/BARD1.
Figure 7. Enhanced nucleosome ubiquitylation activity and binding of full-length BRCA1/BARD1.
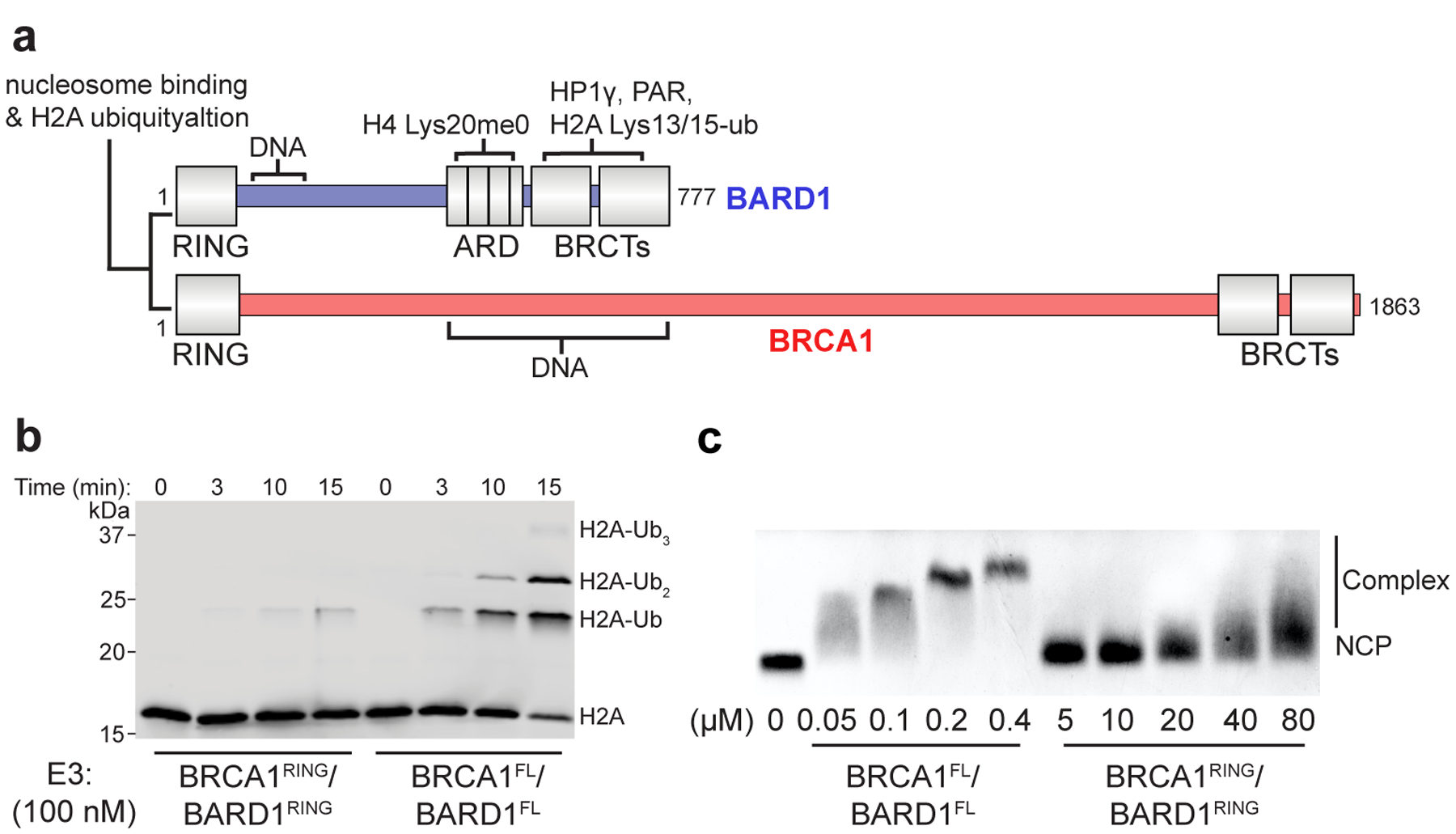
(a) Domain illustration of full-length (FL) BRCA1 and BARD1 with a subset of putative chromatin and protein interactions indicated. (b) Nucleosome ubiquitylation assay comparing the activity of the RING heterodimer (BRCA1RING/BARD1RING) to full-length BRCA1/BARD1 (BRCA1FL/BARD1FL). The E3 concentration was lowered to 100 nM to observe ubiquitylation kinetics of BRCA1FL/BARD1FL. (c) EMSA assay comparing nucleosome binding of BRCA1FL/BARD1FL to BRCA1RING/BARD1RING. The BRCA1/BARD1 concentration used in the binding experiments is reported under the gels and is ~100-fold lower for the full-length construct. Ubiquitylation activity and binding assays are representative of n=2 independent experiments. Uncropped gels in panels b and c are available as source data.
Discussion
We used cryo-EM, NMR, and biochemical methods to reveal the mechanism of site-specific nucleosomal H2A ubiquitylation by BRCA1/BARD1. Our structure shows that while the BRCA1 RING binds to the nucleosome acidic patch using an arginine anchor motif nearly identical to Ring1b and other nucleosome binding factors, BARD1 forms a distinct and novel interface with the H2B/H4 cleft that constrains the BARD1 RING closer to the histone surface than Bmi1 and causes an upward tilt of the BRCA1 E2 binding interface. The E3-E2 complex acts like a teeter-totter, elevating UbcH5c from the histone surface and distancing its active site away from H2A Lys119. This positioning disfavors Ub transfer to Lys119 and promotes modification of lysines 125/127/129 in the disordered tail of H2A that have adequate reach distance from the histone surface to gain access to the UbcH5c active site in the complex. Another potential determinant of specificity suggested from NMR experiments is that the two heterodimeric E3-E2 complexes differentially restrict the conformation of the H2A C-terminal tail. The Ring1b-UbcH5c/Bmi1 complex, positioned closer to the nucleosome surface and the emerging H2A tail, limits the conformational space that can be explored by the C-terminal tail of H2A. This may inhibit H2A lysine residues 125/127/129 from approaching the Ring1b-bound UbcH5c active site and actively prevent off-target modification by Ring1b/Bmi1. Conversely, the retention of native-like flexibility in the BRCA1-UbcH5c/BARD1/nucleosome complex ensures that Lys 125/127/129 are able to approach the E2 active site for Ub transfer. Currently, there is little data regarding the mechanism of substrate lysine approach to E2s, and especially in the case of specific mono-ubiquitylation of disordered substrate lysines. To our knowledge, all reported cases involve substrate lysine residues that are in more ordered protein regions and are observed to be positioned or stabilized near the E2~Ub active site by specific interactions between the E3/substrate47–49 or E2~Ub/substrate50 complexes, and therefore are more similar to the mechanism of Ring1b/Bmi1 and RNF168 than BRCA1/BARD1.
In the structure, UbcH5c binds to the canonical E2 recognition surface of BRCA1. However, the quality of cryo-EM density map for UbcH5c is better adjacent to the BRCA1 subunit and diminishes toward the C-terminal end of the E2. This suggests that the E3/E2 interface is not rigidly fixed, consistent with the moderate affinity of the BRCA1-UbcH5c interaction29. Upon binding, RING E3s, especially those that function with UbcH5c, generally promote a closed conformation of the E2~Ub conjugate to enhance Ub transfer to lysine side chains51. As noted for the Ring1b/Bmi127 and RNF16828 E3-E2/nucleosome complexes, the BRCA1-UbcH5c/BARD1/nucleosome complex is completely compatible with closed conformations of the E2~Ub conjugate (Extended Data Fig. 10). Indeed, tilting the UbcH5c active site away from the nucleosome surface in the BRCA1/BARD1/nucleosome complex would further remove steric restrictions that might impede formation of the closed E2~Ub conformation.
While site-specific BRCA1/BARD1-dependent nucleosome ubiquitylation has been directly observed in vitro for canonical H2A from multiple species and macroH2A, other specialized H2A isoforms including the DNA-damage specific H2Ax that co-localize with BRCA1/BARD1 at DNA DSB foci52 have lysine residues that are predicted to be efficiently ubiquitylated by BRCA1/BARD1 based on our model. Ubiquitylation of these different H2A isoforms in chromatin may delineate its role in various nuclear processes. Although BRCA1/BARD1 can multi-monoubiquitylate the tail of H2A in vitro, a single Ub genetically fused to the C-terminus of H2A has been observed to complement defective BRCA1/BARD1 E3 ligase activity in DNA DSB repair11 and transcriptional regulation15,17. Furthermore, the DUB USP48 was discovered to preferentially target the multi-ubiquitylated H2A-Ub product of BRCA1/BARD1 over Lys13/15-Ub or Lys119-Ub in vitro and to antagonize BRCA1 E3-ligase dependent activity at DSBs in cells18. Further investigation is needed to better understand the biologically relevant H2A-Ub product of BRCA1/BARD1 in vivo and its signaling mechanisms. We envision that downstream chromatin factors will be specifically recruited to chromatin ubiquitylated by BRCA1/BARD1 in a manner similar to the PRC1/253 and the RNF168/53BP121,22 systems.
While our data explain the basis for site-specific nucleosome ubiquitylation by the BRCA1/BARD1 RING heterodimer, we show that the full-length protein has enhanced binding and activity towards a nucleosome substrate. Other regions of BRCA1 and BARD1 are implicated in binding to DNA45,46, histone tails43, and histone modifications44. BRCA1/BARD1 also exists as part of several discrete higher-order molecular assemblies54, including with proteins known to specifically bind to nucleosomes such as PALB255. Our findings suggest that patient mutations in regions critical for these interactions could be pathogenic by indirectly affecting the nucleosome-specific E3 ligase activity of BRCA1/BARD1. Further investigation is needed to understand the chromatin landscape that recruits BRCA1/BARD1 to bind to nucleosomes to ubiquitylate H2A, and which BRCA1/BARD1 complexes are involved in these processes. Finally, our observation that a prevalent BARD1 RING mutation found in cancer patients though not reported to be a causative agent of cancer specifically and potently disrupts H2A ubiquitylation supports the mounting evidence that BRCA1/BARD1-dependent nucleosome ubiquitylation is critical to its function as a tumor suppressor.
Methods
DNA manipulation and protein purification
The BRCA1(1–104)-(GS)n-UbcH5c(2–147) E3-E2 chimeras were constructed using splicing by overlap extension (SOE) PCR56. The GS-linker and an N-terminal precision protease (H3C) cleavage site were added by incorporation into primer sequences. The assembled fragment was sub-cloned into a pCOT7n expression vector downstream of 6xHis tag. After comparison of linker length by EMSA, a construct with sequence BRCA1(1–104)-GSGSGSG-UbcH5c(2–147)/BARD1(26–140) with either Cys85 or Cys85Lys UbcH5c active site was selected for all subsequent biochemical, cryo-EM, and NMR experiments. An H3C cleavage site was also added onto the BRCA1(1–112) construct and cloned into the same pCOT7n expression vector. Untagged human H2A type-2A was inserted into pTEV19 to make the 6xHis-TEV-H2A construct. A VSV-G epitope tag was introduced on a primer to make the 6xHis-TEV-VSV-G-H2A construct. UbcH5c(2–147) was sub-cloned into a modified pET28 vector downstream of a 6xHis-SUMO tag. All mutants were generated by QuikChange site-directed mutagenesis (Agilent) or SOE PCR and sub-cloning. All constructs were verified by Sanger sequencing. Human Uba1 (E1), UbcH5a, UbcH5b, UbcH5c (untagged), Ube2e1, Ube2e2, Ube2e3, ubiquitin, and BRCA1(1–112)/BARD1(26–140) were purified as previously described29,57,58. GST-RNF168(1–189) was purified as previously described without cleaving off the GST tag20. All BRCA1/BARD1 RING heterodimer constructs were expressed by co-transformation of two plasmids containing BRCA1 (pCOT7n vector) and BARD1 (pET28n vector). For BRCA1(1–112)/BARD1(26–140) used in cryo-EM, ITC, and NMR experiments, an additional step was added to remove the 6xHis-tag on BRCA1 with GST-H3C protease on-column overnight as described for the BRCA1-UbcH5c/BARD1 E3-E2 chimera in detail below. The cleaved product was concentrated before a final SEC step using a Superdex 75 column equilibrated in 20 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT on an Akta FPLC (GE Healthcare). Full-length BRCA1/BARD1 was co-expressed using the Bac-to-Bac expression system in Hi5 insect cells and purified as previously described in extensive detail46.
For E2 and E3 expression, Escherichia coli (BL21 DE3) cultures were grown to an optical density at 600 nm (OD600nm) of 0.6–0.8 in LB or MOPS media supplemented with 15N-NH4Cl for NMR isotopic labelling (Cambridge Isotope Labs). Media was supplemented with 100 μM ZnCl2 for RING constructs. Protein expression was induced with 0.2 mM IPTG at 16 °C for 16–18 hours. Bacterial pellets were harvested by centrifugation, resuspended in lysis buffer (25 mM Tris-HCl pH 7.6, 500 mM NaCl, 10 mM imidazole) supplemented with SIGMAFAST EDTA-free protease inhibitor cocktail (Sigma) and 1 mM PMSF, lysed by 2 passages through a French pressure cell, and clarified by centrifugation at 17,000xg at 4 °C using a Sorvall LYNX 6000 centrifuge (Thermo Fisher). The supernatants containing 6xHis-SUMO-UbcH5c or 6xHis-H3C-BRCA1-UbcH5c/BARD1 were bound to a 5 mL HisTrap Ni2+-NTA column (GE Healthcare). After extensive washing with lysis buffer containing 50 mM imidazole, proteins were cleaved on-column using GST-SENP1 or GST-H3C protease (prepared in-house) overnight at 4 °C in 25 mM Tris-HCl pH 7.6, 200 mM NaCl, 10 mM imidazole, 1 mM DTT supplemented with protease inhibitors leupeptin, pepstatin A, and aprotinin (1 μg/mL each). Cleaved protein was washed off the column, concentrated, and purified by SEC using a Superdex 75 column (GE Healthcare) equilibrated in 25 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 1 mM DTT (BRCA1-UbcH5c/BARD1) or 25 mM phosphate pH 7.0, 150 mM NaCl (UbcH5c). To purify Ring1b/Bmi1 constructs (gift from Dr. Song Tan), bacterial supernatants containing STR-10xHis-TEV-Ring1b(2–116)/Bmi1(2–109) or STR-10xHis-TEV-Ring1b(2–116)-GSGSRS-UbcH5c(2–147)/Bmi1(2–109) were bound to a 5 mL HisTrap Ni2+-NTA column (GE Healthcare) in lysis buffer, washed with buffer containing 50 mM imidazole, and eluted in buffer containing 300 mM imidazole. Affinity tags were removed by incubating with 6xHis-TEV protease (prepared in-house) overnight at 4 °C in TEV cleavage buffer (20 mM Tris pH 7.4, 10 μM ZnCl2, 200 mM NaCl, 5 mM citrate, 2 mM 2-mercaptoethanol, 10 mM imidazole). Cleaved product was filtered over Ni2+-NTA resin to re-capture the affinity tag, 6xHis-TEV, and impurities. The flow-through was concentrated and further purified using a Superdex 75 column (GE Healthcare) equilibrated in 25 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 1 mM DTT.
Wild-type and mutant human histones 6xHis-TEV-H2A type-2A (± N-terminal VSV-G tag), untagged H2A type-2A, H2B type-1K, H3.2 (C110A), and H4 were expressed separately in E. coli (BL21 or BL21 pLysS) to an OD600nm of 0.6–0.8 in LB or M9 minimal media and induced with 0.5 mM IPTG for 3 hours (LB) or overnight (M9) at 37 °C. For NMR, M9 media was made in deuterium oxide (D2O) and supplemented with 15N-NH4Cl and fully deuterated 12C-D7-glucose or 13C-D7-glucose to generate perdeuterated histone protein (Cambridge Isotope Labs). Histone octamer formation and purification was performed using the one-pot refolding method59. Briefly, E. coli cell pellets containing all four histones (including 6xHis-TEV-H2A) were mixed together, lysed by sonication in lysis buffer (20 mM Tris-HCl pH 7.5, 500 mM NaCl, 1 mM DTT) then centrifuged (17,000xg, 4 °C) for 30 minutes. The pellet was washed once with lysis buffer, resuspended in extraction buffer (sodium acetate pH 5.2, 7M guanidinium-HCl, 5 mM TCEP) and insoluble debris were removed by centrifugation. Octamers were refolded by dialysis in refolding buffer (20 mM Tris-HCl pH 7.5, 2M NaCl, 10 mM Imidazole, 2 mM ß-mercaptoethanol). After centrifugation, the supernatant containing octamers was purified using a 5 mL HisTrap Ni2+-NTA column in refolding buffer according to manufacturer’s instructions (GE healthcare). The 6xHis-tag was removed using TEV protease (produced in-house) overnight at 4 °C in refolding buffer without imidazole. Octamers were further purified using a Superdex 200 column in 20 mM Tris-HCl pH 7.5, 2 M NaCl, 1 mM DTT (GE Healthcare). For assays with H3 K79 methylation mimetic nucleosomes, wild-type and H3 K79C histones were individually purified using well-established protocols60,61. H3 K79C methylation mimetics were generated as previously described62. Assembled octamers were flash frozen and stored at −80 °C in 20 mM Tris-HCl pH 7.5, 2 M NaCl, 1 mM DTT. Large-scale 153 base-pair Widom 601 DNA (gift from Dr. Lewis Kay) purification was performed essentially as described63 using a plasmid containing 32 tandem repeats separated by EcoRV sites, and NCPs were reconstituted using the standard salt-dialysis method63. As a final step, NCPs were purified by SEC using a Superdex 200 increase 10/300 column (GE Healthcare) equilibrated with 20 mM HEPES-NaOH pH 7.5, 10–50 mM NaCl, 0.01% NaN3, concentrated, and stored on ice for up to two weeks. NCP concentrations were determined by dilution in 0.25 M NaOH and measurement of ultraviolet absorbance at 260 nm using the extinction coefficient 2,866,600 M−1cm−1 to quantify the DNA component. Protein concentrations were determined by their ultraviolet absorbance at 280 nm and confirmed with Coomassie-stained SDS-PAGE.
Electrophoretic mobility shift assays (EMSA)
For EMSA assays in Figs. 1 and Extended Data Fig. 1, 50 nM NCPs were incubated with indicated amount of BRCA1/BARD1 or BRCA1-UbcH5c/BARD1 for 20 mins on ice in 10 mM Tris-HCl pH 7.5, 75 mM NaCl, 1 mM DTT. Sucrose was added to a final concentration of 6%, and samples were run on a pre-chilled 5% polyacrylamide 0.5X TBE gel (40 mM Tris pH 8.3, 45 mM boric acid, 1 mM EDTA) at 140 V for 45 minutes at 4°C. Gels were stained with gel-red dye (Biotium) and visualized using a BioRad gel dock. For assays including full-length BRCA1/BARD1 (Fig. 7), 50 nM NCP were incubated with the indicated amount of BRCA1FL/BARD1FL or BRCA1RING/BARD1RING in 10 μl of buffer (10 mM Tris-HCl, pH 7.5, 0.125 mM DTT, 0.0025% Igepal, 70 mM KCl) on ice for 20 mins. After the addition of 2 μl of gel loading buffer (50% glycerol, 20 mM Tris-HCl, pH 7.4, and 0.05% orange G), NCP complexes were resolved by 1% agarose gel electrophoresis in TAE buffer (40 mM Tris, 20 mM Acetate and 1 mM EDTA) at 4°C with 140 V for 30 mins. The gels were stained in fresh ethidium bromide solution and imaged using a ChemiDocTM MP Imaging System.
SEC-MALS and Analytical SEC binding assays
For SEC-MALS, NCPs (15 μM) were incubated with or without BRCA1-UbcH5cC85K/BARD1 (45 μM) in 20 mM HEPES-NaOH pH 7.5, 35 mM NaCl, 0.5 mM TCEP and injected onto a Agilent HPLC with a Sepax SRT-SEC column equilibrated in the same buffer coupled to a Wyatt miniDAWN TREOS and Optilab T-rEX differential refractive index detector. Molar mass was calculated from the Raleigh ratio based on multi-angle (static) light scattering and protein concentration from the change in refractive index (dn/dc = 0.185). Analysis was performed using Wyatt Astra 7 software. For SEC binding experiments, 10 μM NCPs were incubated with 30 μM of the indicated BRCA1-UbcH5c/BARD1 construct at room temperature for 30 mins in 300 μL binding buffer (20 mM HEPES-NaOH pH 7.5, 25–100 mM NaCl, 0.5 mM TCEP). Samples were then injected onto a Superdex 200 increase (10/300) column equilibrated in binding buffer with the indicated amount of NaCl (25–100 mM) using an Akta FPLC system (GE Healthcare).
Nucleosome ubiquitylation assays
Unless explicitly noted in figure legends, NCP ubiquitylation assays were performed in a reaction mixture containing 0.2 μM E1 (Uba1), 1 μM E2 (UbcH5c), 2 μM E3, 20 μM ubiquitin, and 0.5 μM NCPs in 25 mM HEPES-NaOH pH 7.5, 150 mM NaCl. Several assays (Extended Data Figs 1e and 6b) were performed in a reaction mixture containing 1 μM E1 (Uba1), 4 μM E2 (UbcH5c), 8 μM E3, 20 μM ubiquitin, and 0.5 μM NCPs in 25 mM phosphate pH 7.0, 150 mM NaCl. Unless noted, reactions were performed with wild-type and mutant BRCA11−112/BARD126−140 (referred to as RING heterodimer or BRCA1RING/BARD1RING) as the E3. After collecting zero time-point samples, reactions were brought to 37 °C and 5 mM MgCl2/ATP were added. Reactions were quenched at indicated time points by 1:1 dilution with 4x reducing SDS loading buffer and analyzed by Western blot using rabbit anti-VSV-G (Sigma, V4888), mouse anti-H2A (EMD Millipore, 07–146), mouse anti-H2B (BioRad, VPA00384), rabbit anti-H3 (Abcam, ab1791), or rabbit anti-H4 (Abcam, ab31830) antibodies. Blots were incubated with fluorophore-conjugated anti-mouse (Invitrogen, A21058) or anti-rabbit (Cell Signaling, 5151S) secondary antibodies and visualized using a Licor Odyssey scanner. Quantification was performed using Image Studio software (Licor) by drawing a box measuring the total band intensity for unmodified H2A at each time-point, subtracting background signal, and normalizing to the zero time-point of each reaction.
E2~Ub formation and lysine reactivity assays
For E2 lysine discharge assays, E2~Ub was formed in a reaction consisting of 0.3 μM E1 (Uba1), 60 μM UbcH5c, 120 μM Ubiquitin, 10 mM MgCl2/ATP in 25 mM sodium phosphate, pH 7.0, 150 mM NaCl (PBS) at 37 °C for 30 minutes. The reaction was quenched by addition of 50 mM EDTA, pH 7.4 for 10 minutes, and the lysine discharge reaction was initiated by a 1:1 dilution of the charging reaction into PBS supplemented with 50 mM lysine with or without 3 μM of the indicated E3 at 30 °C. Time-points were taken by mixing the reaction 1:1 with 4x load dye without reducing agent, resolved on an SDS-PAGE gel and stained with Coomassie blue, and visualized on a BioRad gel dock. Quantification was performed in ImageJ (NIH) by drawing a box around the E2~Ub conjugate and measuring the total area of signal and normalizing to the zero time-point. E2 charging experiments were performed using a reaction mixture of 40 μM UbcH5c or BRCA1-UbcH5c/BARD1 (WT or Cys85Lys active site), 2 μM E1 (Uba1), 80 μM Ub, and 10 mM MgCl2/ATP in 50 mM sodium phosphate pH 7.0, 150 mM NaCl (UbcH5c WT active site) or 50 mM CAPS pH 10, 150 mM NaCl (UbcH5c Cys85Lys active site). Reactions were quenched at indicated time points by 1:1 dilution into 4x SDS-PAGE loading dye (± β-mercaptoethanol) and analyzed by Coomassie stained SDS-PAGE gel.
Isothermal Titration Calorimetry (ITC)
For ITC analysis, all proteins were extensively buffer exchanged by dialysis into 20 mM HEPES-NaOH pH 7.5, 50 mM NaCl, 0.5 mM TCEP. Measurements were performed on a MicroCal PEAQ-ITC (Malvern) at 25 °C spanning 19 injections with a stirring speed of 750 rpm. The cell contained 20 μM NCPs and the syringe contained ~800 μM of indicated ligand. Control experiments where ligand was injected into buffer were performed. Binding isotherms were fit to a one-site binding model and figures formatted in the Malvern PEAQ-ITC analysis software package.
Cryo-EM sample preparation, image acquisition, data processing, and model building
Extensive methods related to cryo-EM structure determination can be found as a supplementary note online.
Molecular graphics and structural and sequence alignments
All figures depicting molecular structures were prepared using UCSF Chimera64 or ChimeraX65 software. Alignments were performed using the MatchMaker function in Chimera by best aligning chain, specific chains, or specified residues depending on the analysis. BRCA1/BARD1 and Ring1b/Bmi1 sequence alignments were performed using BLAST66 (NCBI). For structure based alignment of H2A variants, atomic models of nucleosomes containing canonical H2A (PDB id: 5B0Z), macroH2A1 (1U35), H2A.J (6KVD), H2A.X (6K1J), H2A.V (3WAA), H2A.Z (5B33) were aligned and inspected to confirm overlap in the position equivalent to Lys118 from canonical H2A.
NMR sample preparation and data collection
All NMR experiments were collected on an Avance 600 MHz spectrometer equipped with a cryoprobe. Data were processed with the NMRpipe67 package and visualized in NMRviewJ68. All spectra of labelled NCP samples were collected at 25 °C in 20 mM MOPS pH 7.0, 50 mM NaCl, 0.5 mM TCEP in the presence of 10% D2O. Spectra of BRCA1/BARD1 RING heterodimers were collected at 30 °C in 25 mM phosphate pH 7.0, 150 mM NaCl, 10% D2O. To assign the H2A tails in the NCP, a standard set of multidimensional TROSY NMR experiments (HNCA, HNCOCA, HNCACB) were used. Triple-resonance experiments were performed with 180 μM 2H15N13C-H2A NCPs where all other histones and DNA contained natural abundance isotopes. The H2A construct for NMR contained a short, non-native N-terminal sequence after TEV cleavage (GAMG). Assignments were performed by correlating peaks among the triple resonance experiments in NMRViewJ.
For 1H15N-TROSY-HSQC NMR binding experiments, concentrated NCPs were buffer exchanged into 20 mM MOPS pH 7.0, 10 mM NaCl, 0.5 mM TCEP. BRCA1-UbcH5c/BARD1 and Ring1b-UbcH5c/Bmi1 constructs were buffer exchanged into 20 mM MOPS pH 7.0, 150 mM NaCl, 0.5 mM TCEP to prevent self-association in the absence of NCP binding partner in low-salt. NMR samples containing 15 μM 2H15N-H2A NCPs were assembled by stepwise addition of E3-E2 binding partner over ~30 minutes until saturation of the complex. The final salt concentration of all NMR samples was adjusted to 50 mM NaCl and supplemented with 10% D2O. For the Ring1b-UbcH5c/Bmi1/NCP complex, irreversible aggregation was observed immediately after saturation (30 μM), resulting in minimal sample loss. BRCA1-UbcH5cC85K/BARD1 did not aggregate with the NCP and was added to a final concentration of 60 μM (or more) to account for its weaker binding affinity. NMR samples with 2H15N-H3 NCPs were assembled identically to H2A-labelled samples. Resonance assignments for the N-terminal tail of H3 were directly transferred from a previous study69. Peak intensities were measured using NMRViewJ, and the intensity of resonances in the bound complex spectrum were compared to those in the apo reference spectrum (Icom/Iref).
Reporting Summary Statement
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data Availability Statement
The cryo-EM map of the BRCA1-UbcH5c/BARD1/nucleosome complex was deposited to the Electron Microscopy Data Bank under accession code EMD-22581 and the atomic model to the Protein Data Bank under accession code 7JZV. NMR chemical shift assignments were deposited to the Biological Magnetic Resonance Data bank under accession code 50604. Uncropped gels/blots and statistical source data are available online linked to this article. Plasmid reagents generated in this study can be obtained upon request from the corresponding author Dr. Rachel Klevit.
Extended Data
Extended Data Fig. 1. Specificity of BRCA1/BARD1-dependent nucleosome ubiquitylation and validation of the E3-E2 chimera.
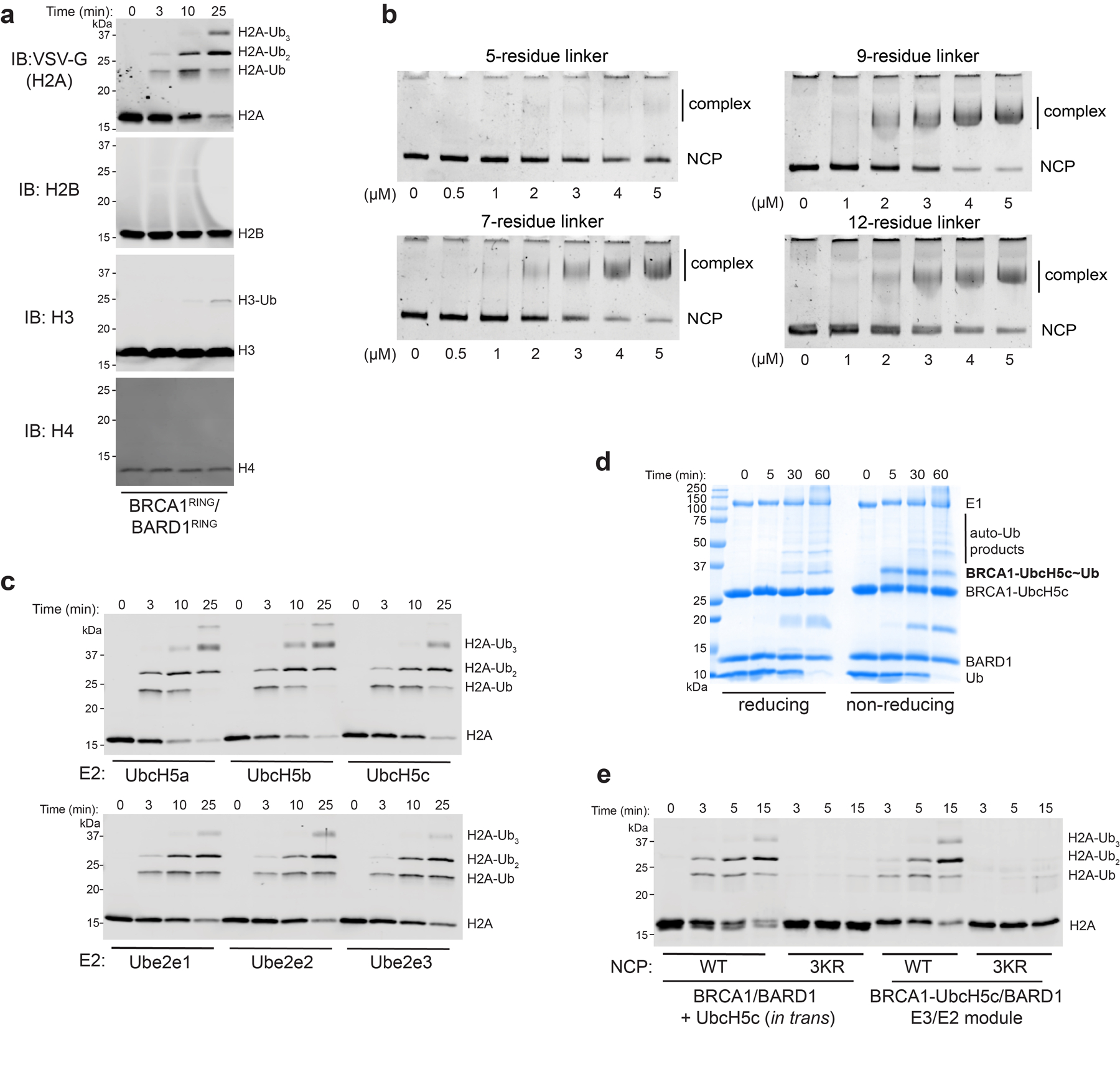
(a) Western blot analysis of all four histone subunits from the same nucleosome ubiquitylation reaction. (b) Native-gel EMSA measuring NCP binding of BRCA1-UbcH5c/BARD1 constructs with various Gly/Ser-repeat linker lengths. The E3-E2 chimera with a seven-residue linker was used for structure determination and all subsequent experiments. (c) Nucleosome ubiquitylation assays using BRCA1/BARD1 with indicated E2 enzymes. (d) Coomassie-stained gel under reducing or non-reducing conditions of an E2 charging reaction using the BRCA1-UbcH5c/BARD1 chimera. (e) Nucleosome ubiquitylation activity of BRCA1/BARD1 with UbcH5c in trans or the BRCA1-UbcH5c/BARD1 chimera with wild-type (WT) or Lys125/127/129Arg (3KR) NCP substrates. Data in panels c and e are representative of n=2 independent experiments. Uncropped gels/blots in panels a-e are available as source data.
Extended Data Fig. 2. Purification of a stable BRCA1-UbcH5c/BARD1/nucleosome complex.
(a) Coomassie-stained gel of an E2 charging reaction using UbcH5cC85K and BRCA1-UbcH5cC85K/BARD1. (b) Size exclusion chromatography (SEC) of NCPs (black) with excess BRCA1-UbcH5c/BARD1 (purple) or BRCA1-UbcH5cC85K/BARD1 (green) with 35 mM NaCl in SEC buffer. (c) Coomassie-stained gels of fractions from SEC binding experiments shown in panel b. (d) SEC of NCPs with excess BRCA1-UbcH5cC85K/BARD1 with 100 mM NaCl (black) or 25 mM NaCl (green) in SEC running buffer. (e) Coomassie-stained gels of fractions from SEC binding experiments shown in panel d. (f) Size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) analysis of NCPs (black) and the BRCA1-UbcH5cC85K/BARD1/NCP complex (green). Dashed lines report MALS molecular weight (MW) data. The MW value reported is the average MW from light scattering ± 1-s.d. where error is a measure of statistical consistency of light scattering data and not an absolute bound on the error of MW. The expected MW is 203.7 kDa for the NCP and 287.2 kDa for the complex bound with 2:1 stoichiometry. Uncropped gels in panels a, c, and e are available as source data.
Extended Data Fig. 3. Cryo-EM processing workflow.
(a) Flow chart of data processing steps. Objects with dark borders represent solvent masks and are not density maps. (b) Representative motion-corrected micrograph from data set. White scale bar is 100 nm.
Extended Data Fig. 4. Cryo-EM validation and example density.
(a) Local resolution estimate from CryoSparc after non-uniform refinement at the FSC=0.143 cutoff plotted on the final density modified map of the whole complex (left panel) and close-up on the RING-histone interface (right panel). (b) Half-map Fourier shell correlation curves from CryoSparc non-uniform refinement (red) and after density modification (blue). (c) Euler angle distribution of refined particle subset used in final cryo-EM reconstruction. (d) Main chain trace of the BRCA1 RING, BARD1 RING, four-helix bundle, and UbcH5c fit into cryo-EM map. The active site position of UbcH5c (Lys 85) Cα is shown as a sphere. (e) Subset of important histone regions fit into the cryo-EM map with side chains shown (except for H2A C-terminal tail).
Extended Data Fig. 5. Extensive analysis of the BRCA1/BARD1 RING-histone interface.
(a, b, c) Close-up view of BRCA1 and BARD1 RING domains in the complex showing side chains of residues mutated in nucleosome ubiquitylation assays in panels d-g and j-k (d) Representative nucleosome ubiquitylation assay using the indicated BRCA1 RING mutants. (e) Quantified nucleosome ubiquitylation assays for BRCA1 mutants shown in panel d. (f, g) Same experimental set-up as panels d and e using the indicated BARD1 mutants. (h) ITC binding data for BRCA1/BARD1 and indicated mutants with NCPs. Summarized data are also shown in figure 3e. (i) 1H15N-TROSY-HSQC spectra of wild-type (WT) BRCA1/BARD1 and BRCA1(WT)/BARD1(Trp91Ala). (j, k) Same experimental set-up as panels d and e using BARD1 mutants reported on the COSMIC database. (l) E2~Ub lysine discharge assay using the indicated BRCA1/BARD1 mutants. (m) Quantified E2~Ub lysine discharge assays using the BRCA1/BARD1 mutants shown in panel l. (n, o) Same experimental set-up as panels l and m using BARD1 COSMIC mutants. Quantified nucleosome ubiquitylation assay data show the mean and error bars are ± 1-s.d. of n=3 independent experiments at 10-minute (gray bars) and 30-minute (colored bars) time points. Statistical analysis is compared to wild-type at the 10-minute time point. Quantified E2 lysine discharge data show the mean and error bars are ± 1-s.d. of n=3 independent experiments at 2-, 6- and 10- minute time points (light, medium, and dark gray bars). Statistical comparisons are indicated with lines above the graphs. All p-values were calculated using a two-tailed Student’s t-test (* p≤0.05, ** p≤0.005, ns = not significant). Uncropped gels/blots in panels d, f, j, l, and n and data from graphs in panels e, g, k, m, and o are available as source data.
Extended Data Fig. 6. Comparison of nucleosome requirements for H2A-modifying E3s.
(a) Comparison of the conserved BRCA1 and Ring1b arginine anchor motif interactions (PDB: 4R8P). Complexes were aligned by H2B on the bound face of the NCP. H2A from the Ring1b-UbcH5c/Bmi1 nucleosome complex is shown in gray. (b) Nucleosome ubiquitylation assays using the indicated H2A/H2B acidic patch NCP mutants with BRCA1/BARD1 (top) and Ring1b/Bmi1 (bottom). (c) Comparison of interactions between the RING domains of BRCA1/BARD1 and Ring1b/Bmi1 with the H2B αC helix residues assayed in panels d and e. (d) Representative nucleosome ubiquitylation assays using the indicated H2B αC helix NCP mutants with BRCA1/BARD1 (top) or Ring1b/Bmi1 (bottom). (e) Quantified assays using the NCP mutants and E3s from panel d (f, g) Same experimental set-up as in panels d and e using H2B/H4 cleft NCP mutants. (h) BARD1-histone binding interface with side chain density for H3 Phe78 and Lys79 shown as semi-transparent surface. (i, j) Same experimental set-up as panels d and e using H3 Lys79Ala NCP mutant. (k, l, m) Representative nucleosome ubiquitylation assays using H3 Lys79me1 and Lys79me2 mimetic NCP substrates with the indicated E3s. (n, o, p) Quantified assays using H3 Lys79 methylation mimetic nucleosome substrates and the indicated E3s. Quantified ubiquitylation assays show the mean and error bars are ± 1-s.d. of n=3 independent experiments (except panel g where n=6 for wild-type) plotted for 10-minute (gray bars) and 30-minute (colored bars) time points. P-values for each mutant were calculated using a two-tailed Student’s t-test compared to wild-type (* p≤0.05, ** p≤0.005, ns = not significant). Panels g (top), and n are also shown in figure 3. Uncropped blots in panels b, d, f, i, k, l, and m and data from graphs in panels e, g, j, n, o, and p are available as source data.
Extended Data Fig. 7. Comparison to the unbound RING heterodimer and Ring1b-UbcH5c/Bmi1/nucleosome complexes.
(a) Structural alignment of the BRCA1/BARD1 RING heterodimers from the NCP complex and the unbound structure (PDB: 1JM7, NMR ensemble model 1) aligned by the four-helix bundle. (b) Structural alignment of the NCP from the indicated complexes by H2B on the E3-E2-bound side. (c) Structural alignment of BRCA1-UbcH5c and Ring1b-UbcH5c from E3-E2/NCP complexes. (d) Comparison of BRCA1-bound and Ring1b-bound UbcH5c in NCP complexes showing side chains mutated in panels g and h. Some atoms for Arg72 and Lys128 are not modelled in the Ring1b-bound structure. Green spheres are Cα of H2A Lys118. (e) Analysis of NCP binding by the BRCA1/BARD1 RING heterodimer and the BRCA1-UbcH5c/BARD1 (WT UbcH5c active site) chimera by ITC. Data are representative of n=2 experiments. (f) Comparison of NCP binding strength for BRCA1/BARD1 and Ring1b/Bmi1 RING heterodimer and E3-E2 chimeras. Affinities for BRCA1/BARD1 were determined using ITC while affinities for Ring1b/Bmi1 were measured using a fluorescence based binding assay27. (g) Representative nucleosome ubiquitylation assays using BRCA1/BARD1 or Ring1b/Bmi1 with indicated UbcH5c mutants. (h) Quantification of nucleosome ubiquitylation using the UbcH5c mutants and E3s from panel g. Data show the mean and error bars are ± 1-s.d. of n=3 independent experiments plotted for 10-minute (gray bars) and 30-minute (colored bars) time points of the reactions. P-values for each mutant were calculated using a two-tailed Student’s t-test compared to wild-type (* p≤0.05, ** p≤0.005, ns = not significant). Uncropped blots in panel g and data from graphs in panel h are available as source data.
Extended Data Fig. 8. NMR analysis of E3-E2/nucleosome complexes.
(a) 1H15N-TROSY-HSQC spectrum of 2H15N13C-H2A in NCP (~180 μM sample) with residue assignments plotted. Signals with asterisks were not able to be assigned. Residues 120# and 120## appear to be alternate confirmations of 120 based on Cα chemical shifts of residues 119 and 120 in the HNCA and HNCOCA spectra. Assignment of the minor population for 6# was based on the similar back bone H, N, and Cα chemical shifts as compared to the major residue 6 population. (b) Sequence of H2A N- and C-terminal tails observable in NMR spectrum in panel a with assigned residues bolded. (c) Overlay of 1H15N-TROSY-HSQC spectra of 2H15N-H2A in NCP, with BRCA1-UbcH5cC85K/BARD1 added at increasing concentrations. (d) Quantification of signal broadening from spectra in panel c as a function of BRCA1-UbcH5cC85K/BARD1 added, normalized to the first titration point (60 μM addition). Signal behavior clusters into two groups, where resonances from the extreme N- and C- terminal tails lose signal at a similar rate (cluster 1). (e) Overlay of 1H15N-TROSY-HSQC spectra of 2H15N-H2A in NCP (black, bottom), with BRCA1-UbcH5cC85K/BARD1 added (red, middle), or BRCA1-UbcH5c/BARD1 (blue, top). (f) Quantification of intensity for H2A signals from spectra in panel e comparing the bound complexes to the apo reference spectrum (Icom/Iref). (g) Overlay of 1H15N-TROSY-HSQC spectra of 2H15N-H3 in NCP (black, bottom), with BRCA1-UbcH5cC85K/BARD1 added (red, middle), or Ring1b-UbcH5c/Bmi1 (pink, top). (h) Quantification of intensity for H2A signals from spectra in panel g comparing the bound complexes to the apo reference spectrum (Icom/Iref). Concentrations of E3-E2 complexes added as well as labelled NCPs in the experiment are indicated in the figure. Data from graphs in panels d, f, and h are available as source data.
Extended Data Fig. 9. Biochemical purity and H2A specificity of full-length BRCA1/BARD1.
(a) Coomassie-stained gel of purified full-length BRCA1/BARD1. (b) Nucleosome ubiquitylation assay using full-length BRCA1/BARD1 and wild-type (WT) or H2A Lys125/127/129Arg (3KR) nucleosome substrates. Uncropped gels/blots in panels a and b are available as source data.
Extended Data Fig. 10. Compatibility of closed E2~Ub conformations.
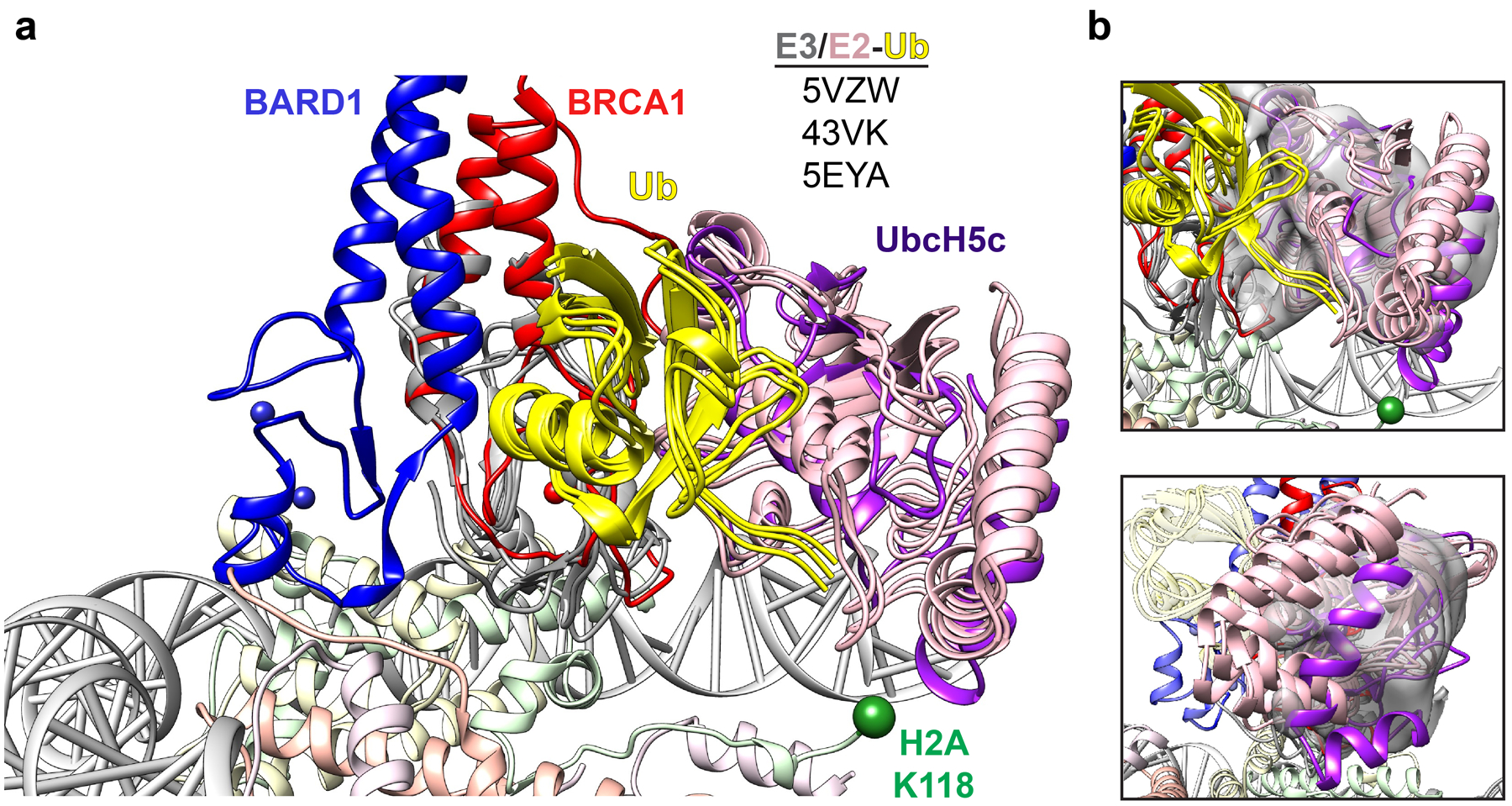
(a) Structural overlay of the BRCA1-UbcH5c/BARD1/nucleosome complex with indicated RING/E2-Ub crystal structures aligned to the RING domain of BRCA1. (b) Comparison of UbcH5c location in the nucleosome complex (purple) and indicated reported RING/E2-Ub structures from panel a with cryo-EM density for UbcH5c shown as gray volume. Density for the E2 is shown from a map that was locally filtered in CryoSparc to decrease excessive noise introduced from map sharpening in this region.
Supplementary Material
Acknowledgements
We thank S. Tan (Penn State University) for sharing Ring1b/Bmi1plasmids and L. Kay (University of Toronto) for sharing the 153 base-pair Widom 601 repeat plasmid. We thank N. Zheng for his insightful feedback on the manuscript and sharing laboratory equipment, T. Hinds for assistance with ITC experiments, P. Hsu for advice on nucleosome complexes, J. Quispe and Q. Beedle for technical assistance with cryo-EM data collection, and L. Walls, A. Borst, and D. Veesler for advice on single-particle cryo-EM data processing. S.R.W. acknowledges support from the NIH (T32GM008268). A.L.B. was supported by the NIH (T32GM008268 and 1F31EY030732), D.P.F. was supported through the Human Frontiers Science Program (RGP0061/2019). J.M.H. was supported by the NIH (T32GM007270). W.Z. was supported by a V Scholar Grant (V2019.Q13) from V Foundation and a Young Investigator Award from Max and Minnie Tomerlin Voelcker Fund. F.D. was supported by the NIH (GM123089). R.E.K. is the Edmond H. Fischer Endowed Chair in Biochemistry and is supported by the NIH (GM088055).
Footnotes
Competing Interests Statement
The authors declare no competing interests.
References
- 1.Tarsounas M & Sung P The antitumorigenic roles of BRCA1–BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol 21, 284–299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Densham RM & Morris JR Moving Mountains—The BRCA1 Promotion of DNA Resection. Front. Mol. Biosci 6, 79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hashizume R et al. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem 276, 14537–14540 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Brzovic PS, Rajagopal P, Hoyt DW, King MC & Klevit RE Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat. Struct. Biol 8, 833–837 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Wu W et al. BRCA1 Ubiquitinates RPB8 in Response to DNA Damage. Cancer Res. 67, 951 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Sato K et al. A DNA-damage selective role for BRCA1 E3 ligase in claspin ubiquitylation, CHK1 activation, and DNA repair. Curr. Biol. CB 22, 1659–1666 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Vázquez-Arreguín K et al. BRCA1 through Its E3 Ligase Activity Regulates the Transcription Factor Oct1 and Carbohydrate Metabolism. Mol. Cancer Res. MCR 16, 439–452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reid LJ et al. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc. Natl. Acad. Sci. U. S. A 105, 20876–20881 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shakya R et al. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 334, 525–528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drost R et al. BRCA1 RING Function Is Essential for Tumor Suppression but Dispensable for Therapy Resistance. Cancer Cell 20, 797–809 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Densham RM et al. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol 23, 647–655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Densham RM & Morris JR The BRCA1 Ubiquitin ligase function sets a new trend for remodelling in DNA repair. Nucleus. 8, 116–125 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thakar A, Parvin JD & Zlatanova J BRCA1/BARD1 E3 Ubiquitin Ligase Can Modify Histones H2A and H2B in the Nucleosome Particle. J. Biomol. Struct. Dyn 27, 399–405 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Kalb R, Mallery DL, Larkin C, Huang JTJ & Hiom K BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep. 8, 999–1005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Q et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477, 179–184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim B-J et al. The Histone Variant MacroH2A1 Is a BRCA1 Ubiquitin Ligase Substrate. Cell Rep. 19, 1758–1766 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart MD et al. BARD1 is necessary for ubiquitylation of nucleosomal histone H2A and for transcriptional regulation of estrogen metabolism genes. Proc. Natl. Acad. Sci. U. S. A 115, 1316–1321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uckelmann M et al. USP48 restrains resection by site-specific cleavage of the BRCA1 ubiquitin mark from H2A. Nat. Commun 9, 229 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattiroli F et al. RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Mattiroli F, Uckelmann M, Sahtoe DD, van Dijk WJ & Sixma TK The nucleosome acidic patch plays a critical role in RNF168-dependent ubiquitination of histone H2A. Nat. Commun 5, 3291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fradet-Turcotte A et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson MD et al. The structural basis of modified nucleosome recognition by 53BP1. Nature 536, 100–103 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Wang H et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Cao R, Tsukada Y & Zhang Y Role of Bmi-1 and Ring1A in H2A Ubiquitylation and Hox Gene Silencing. Mol. Cell 20, 845–854 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Tamburri S et al. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol. Cell 77, 840–856.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vissers JHA, van Lohuizen M & Citterio E The emerging role of Polycomb repressors in the response to DNA damage. J. Cell Sci 125, 3939 (2012). [DOI] [PubMed] [Google Scholar]
- 27.McGinty RK, Henrici RC & Tan S Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horn V et al. Structural basis of specific H2A K13/K15 ubiquitination by RNF168. Nat. Commun 10, 1751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brzovic PS et al. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc. Natl. Acad. Sci. U. S. A 100, 5646–5651 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson CJ et al. Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase. Cell Rep. 26, 1681–1690.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valencia-Sánchez MI et al. Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination. Mol. Cell 74, 1010–1019.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Y et al. High-resolution comparative modeling with RosettaCM. Struct. Lond. Engl 1993 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J et al. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci 117, 1496 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGinty RK & Tan S Recognition of the nucleosome by chromatin factors and enzymes. Curr. Opin. Struct. Biol 37, 54–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taherbhoy AM, Huang OW & Cochran AG BMI1–RING1B is an autoinhibited RING E3 ubiquitin ligase. Nat. Commun 6, 7621 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Wood K, Tellier M & Murphy S DOT1L and H3K79 Methylation in Transcription and Genomic Stability. Biomolecules 8, 11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen AT & Zhang Y The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 25, 1345–1358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huyen Y et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432, 406–411 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Wysocki R et al. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol 25, 8430–8443 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tate JG et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47, D941–D947 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah S, Verma T, Rashid M, Gadewal N & Gupta S Histone H2A isoforms: Potential implications in epigenome plasticity and diseases in eukaryotes. J. Biosci 45, 4 (2020). [PubMed] [Google Scholar]
- 42.Carrion-Vazquez M, Marszalek PE, Oberhauser AF & Fernandez JM Atomic force microscopy captures length phenotypes in single proteins. Proc. Natl. Acad. Sci 96, 11288 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakamura K et al. H4K20me0 recognition by BRCA1–BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol 21, 311–318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becker JR et al. BARD1 links histone H2A Lysine-15 ubiquitination to initiation of BRCA1-dependent homologous recombination. bioRxiv 2020.06.01.127951 (2020) doi: 10.1101/2020.06.01.127951. [DOI] [Google Scholar]
- 45.Masuda T, Xu X, Dimitriadis EK, Lahusen T & Deng C-X ‘DNA Binding Region’ of BRCA1 Affects Genetic Stability through modulating the Intra-S-Phase Checkpoint. Int. J. Biol. Sci 12, 133–143 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao W et al. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature 550, 360–365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Streich FC Jr & Lima CD Capturing a substrate in an activated RING E3/E2-SUMO complex. Nature 536, 304–308 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott DC et al. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 157, 1671–1684 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown NG et al. RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex. Proc. Natl. Acad. Sci 112, 5272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eddins MJ, Carlile CM, Gomez KM, Pickart CM & Wolberger C Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol 13, 915–920 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Pruneda JN et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol. Cell 47, 933–942 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paull TT et al. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. CB 10, 886–895 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Cooper S et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun 7, 13661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savage KI & Harkin DP BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability. FEBS J. 282, 630–646 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Belotserkovskaya R et al. PALB2 chromatin recruitment restores homologous recombination in BRCA1-deficient cells depleted of 53BP1. Nat. Commun 11, 819 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aiyar A, Xiang Y & Leis J Site-directed mutagenesis using overlap extension PCR. Methods Mol. Biol. Clifton NJ 57, 177–191 (1996). [DOI] [PubMed] [Google Scholar]
- 57.Christensen DE, Brzovic PS & Klevit RE E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol 14, 941–948 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Lazar GA, Desjarlais JR & Handel TM De novo design of the hydrophobic core of ubiquitin. Protein Sci. Publ. Protein Soc 6, 1167–1178 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee Y-T, Gibbons G, Lee SY, Nikolovska-Coleska Z & Dou Y One-pot refolding of core histones from bacterial inclusion bodies allows rapid reconstitution of histone octamer. Protein Expr. Purif 110, 89–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luger K, Rechsteiner TJ & Richmond TJ Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 (1999). [DOI] [PubMed] [Google Scholar]
- 61.Dhall A et al. Sumoylated human histone H4 prevents chromatin compaction by inhibiting long-range internucleosomal interactions. J. Biol. Chem 289, 33827–33837 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simon MD et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dyer PN et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Goddard TD et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. Publ. Protein Soc 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul SF, Gish W, Miller W, Myers EW & Lipman DJ Basic local alignment search tool. J. Mol. Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 67.Delaglio F et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 68.Johnson BA Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. Clifton NJ 278, 313–352 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Morrison EA, Bowerman S, Sylvers KL, Wereszczynski J & Musselman CA The conformation of the histone H3 tail inhibits association of the BPTF PHD finger with the nucleosome. eLife 7, e31481 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM map of the BRCA1-UbcH5c/BARD1/nucleosome complex was deposited to the Electron Microscopy Data Bank under accession code EMD-22581 and the atomic model to the Protein Data Bank under accession code 7JZV. NMR chemical shift assignments were deposited to the Biological Magnetic Resonance Data bank under accession code 50604. Uncropped gels/blots and statistical source data are available online linked to this article. Plasmid reagents generated in this study can be obtained upon request from the corresponding author Dr. Rachel Klevit.


