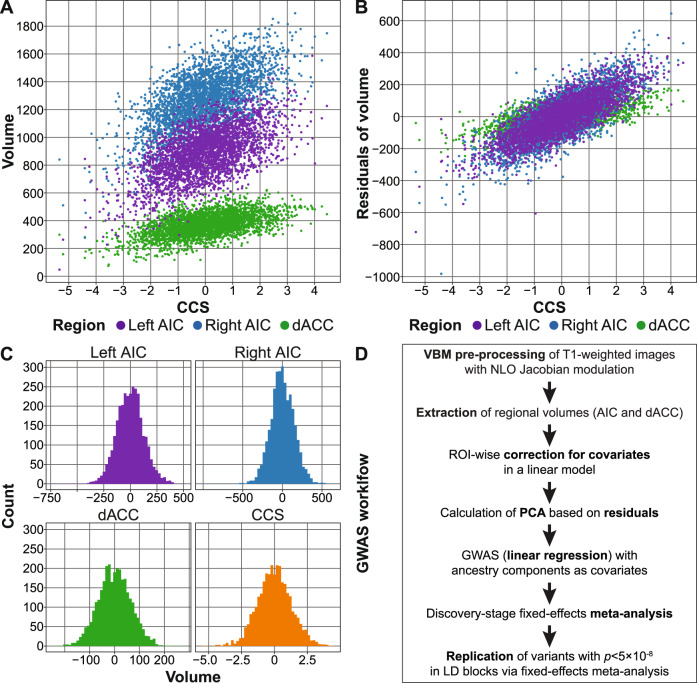
Fig. 1. Generation of the component of the common neurobiological substrate (CCS) and genome-wide association study (GWAS) analysis workflow.
A, B Comparison between the CCS and the three individual volumes (A) and the residuals of the three volumes after correction for covariates (B). AIC anterior insula cortex, dACC dorsal anterior cingulate cortex. C Histograms of the three extracted volumes and the CCS. Note that A–C show combined data from all five GWAS cohorts. Correlations were left and right AIC: r = 0.65, left AIC and dACC: r = 0.52, right AIC and dACC: r = 0.46. D GWAS analysis workflow. All measures were extracted using NLO-based Jacobian modulation. All GM volumes were corrected for age, age2, and sex as covariates; handedness was used as an additional covariate for the three samples 1000BRAINS, CONNECT100, and BiDirect. PCA: principal component analysis, LD: linkage disequilibrium.