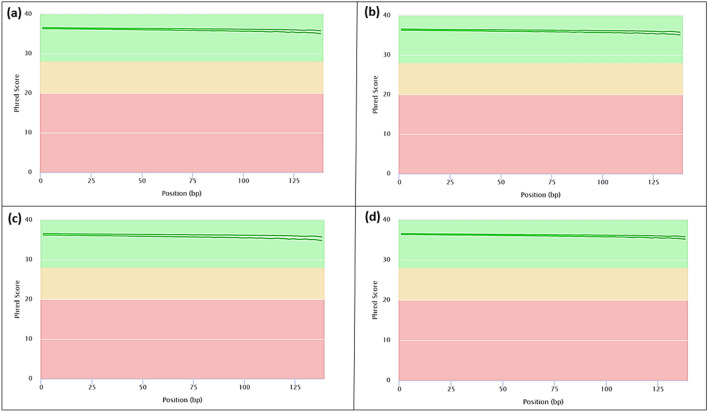
Fig. 3.
Sequence quality of Cinnamon RNA extracted using the modified CTAB RNA extraction protocol. RNA was assessed by producing high-throughput RNA sequencing (RNA-seq) libraries of the different cinnamon tissue samples. The RNA-Seq libraries were pair-end sequenced (150 bp) on the Illumina NovaSeq6000 Platform. Sequencing quality assessment using FastQC version 0.10.1 (Andrews 2014) is represented in graphs describing quality across all bases from every sequence read at cinnamon a Bark; b Leaf; c Flower; d Root tissues. †Phred quality score numerically expresses the accuracy of each nucleotide. Higher Q number signifies higher accuracy. For example, if Phred assigns a quality score of 30 to a base, the chances of having base call error are 1 in 1000. The background of the graph divides the y-axis into very good quality calls (green), reasonable quality (orange), and poor quality (red). The graphs are representative of the forward and reverse reads