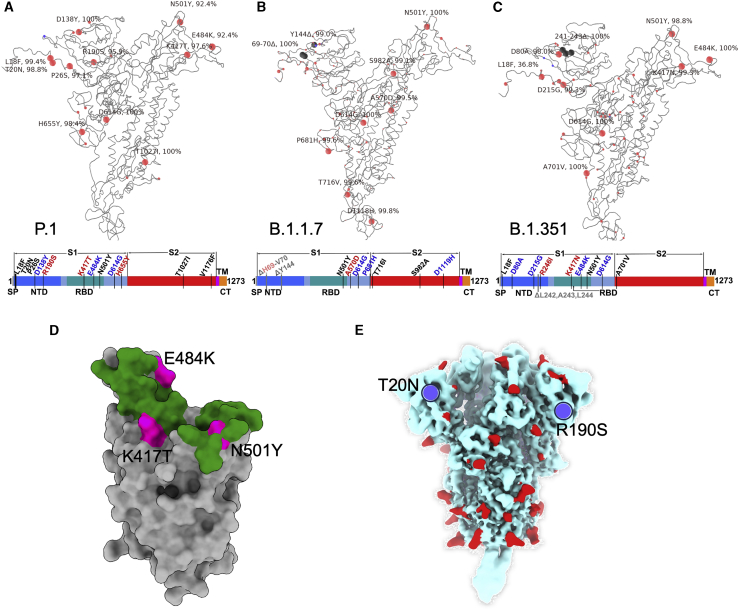
Figure 1.
Mutational landscape of P.1
(A–C) Schematic showing the locations of amino acid substitutions in P.1 (A), B.1.1.7 (B), and B.1.351 (C) relative to the Wuhan SARS-CoV-2 sequence. The time course of P.1 emergence is shown in Figure S1. Point mutations are shown in red and deletions in dark gray. Under the structural cartoon is a linear representation of S with changes marked on. Where there is a charge change introduced by mutations, the change is colored (red if the change makes the mutant more acidic/less basic and blue if the change makes the mutant more basic/less acidic).
(D) Depiction of the RBD as a gray surface with the location of the three mutations (K417T, E484K, and N501Y) (magenta); the ACE2 binding surface of RBD is colored green.
(E) Locations of N-linked glycan (red surface) on the S trimer shown in a pale blue surface representation, and the two new sequons found in P.1 are marked in blue.