Figure 2.
Comparison of wild-type (WT) RBD-ACE2 and P.1 RBD-ACE2 complexes
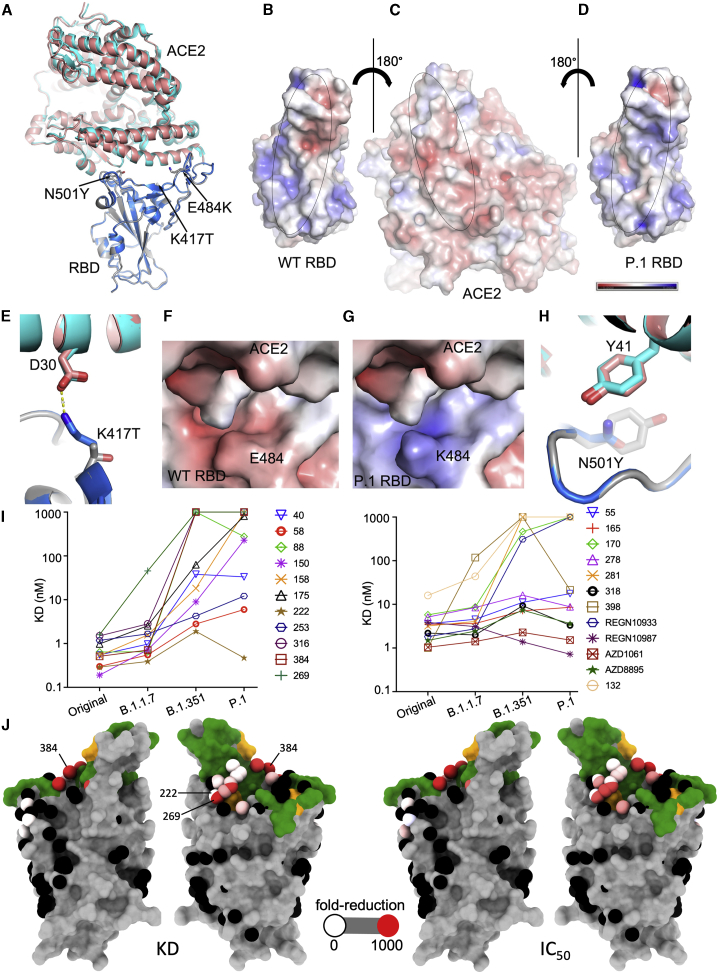
(A) Comparison of P.1 RBD-ACE2 (gray and salmon) with WT RBD-ACE2 (blue and cyan) (PDB: 6LZG) by overlapping the RBDs. The mutations in the P.1 RBD are shown as sticks.
(B–D) Open-book view of electrostatic surface of the WT RBD-ACE2 complex (B) and the P.1 RBD/ACE2 complex (C and D). Note the charge difference between the WT and the mutant RBDs. The charge range displayed is ±5 kJ/mol.
(E) The K417 of the WT RBD forms a salt bridge with D30 of ACE2.
(F and G) Effect of E484K mutation on the electrostatic surface. The tight binding of ACE2 is demonstrated by BLI analysis in Figure S2.
(H) Y501 of the P.1 RBD makes a stacking interaction with Y41 of ACE2.
(I) KD of RBD-mAb interaction measured by BLI for RBDs of Victoria, B.1.1.7, P.1, and B.1.351 (left to right)
(J) BLI data mapped onto the RBD using the method described previously (Dejnirattisai et al., 2021). Front and back views of the RBD are shown. In the left pair, the spheres represent the antibody binding sites colored according to the ratio (KDP.1/KDWuhan). For white, the ratio is 1; for red, it is <0.1 (i.e., at least 10-fold reduction). Black dots refer to mapped antibodies not included in this analysis. Dark green indicates the RBD ACE2 binding surface. Yellow marks mutated K417T, E484K, and N501Y. For the right pair, spheres are colored according to the log of the ratio of neutralization titers (IC50P.1P.1/IC50Victoria). For white, the ratio is 1; for red, it is <0.001 (i.e., at least 1,000-fold reduction). Note the strong agreement between KD and IC50. All relevant data are shown in Table S2.